Unraveling the Success of ADA and PDK1 Overexpressed CAR T cells in Clearing Colorectal Adenocarcinoma Through RNA Sequencing Analysis
Grade 9
Presentation
No video provided
Problem
Chimeric antigen receptor (CAR) T cell therapy has been widely successful in treating patients with hematological malignancies, due to their homogenous tumor nature allowing a single antigen to be targeted by CAR-T cells (Sterner & Sterner, 2021). Conversely, solid tumors, like those formed with colorectal cancer, have heterogenous phenotypes, genetic material profiles, and proteins among the affected area and patients themselves, making it difficult to identify a single target antigen, and prevent immunosuppression within the metabolically unfavorable tumor microenvironment (Johnson et al., 2022). Colorectal cancer is the third most common cancer globally, predominantly affecting individuals 50 and older due to the extensive growth time of colorectal polyps (World Health Organization, 2023). However, colorectal cancer incidence has significantly increased for Canadians under 50 (O’Sullivan et al. (2020), underscoring the importance of discovering effective treatment.
Current limitations in CAR T-cell therapy efficiency include:
-
Antigen Heterogeneity: Some tumors exhibit variability in the expression of target antigens. This means that not all cancer cells within a tumor may express the same antigen targeted by the CAR T cells. As a result, CAR T cells may not effectively recognize and eliminate all cancer cells, leading to treatment resistance or relapse.
-
T Cell Exhaustion: CAR T cells may become exhausted due to prolonged exposure to tumor antigens and the tumor microenvironment. Exhausted T cells lose their effector functions, become dysfunctional, and fail to adequately proliferate and target cancer cells. This exhaustion can limit the effectiveness of CAR T cell therapy and contribute to treatment failure.
-
Development of Resistance: Cancer cells can develop mechanisms to evade CAR T cell therapy, leading to treatment resistance and disease relapse. This resistance can occur through various mechanisms, including downregulation or mutation of target antigens, alterations in tumor signaling pathways, and immune evasion strategies. This can also occur from T cell dysfunction and exhaustion.
-
Antigen Escape: A pathogen’s binding sites typically become more similar to its hosts immune cells over time, causing CAR T-cells to become unable to recognize tumor-specific cells and begin attacking naturally occurring lymphocytes.
With little research into the genomic aspect of efficient CAR T-cells in colorectal cancer clearance, and its historical incidence increase, it is imperative to discover the potential of CAR T-cells in clearing colorectal cancer adenocarcinoma.
This bioinformatic project identifies novel indicators of successful CAR T-cell clearance in solid tumors (specifically colorectal cancer), through RNA sequencing analysis of 3-phosphoinositide-dependent kinase 1 (PDK1) and Adenosine Deaminase (ADA) overexpressed CAR T-cells.
Objectives:
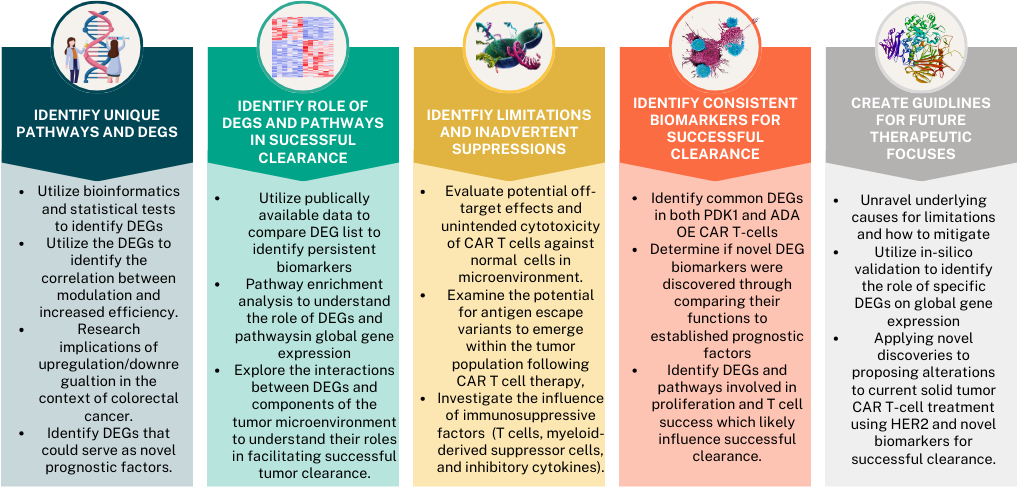
The guiding question is:
What molecular, biological, cellular, KEGG pathways, and gene suppression and overexpression, are involved in successful CAR T-cell clearance of colorectal cancer, and how can identifying these biomarkers mitigate current limitations in CAR T-cell clearance of solid tumors?
Method
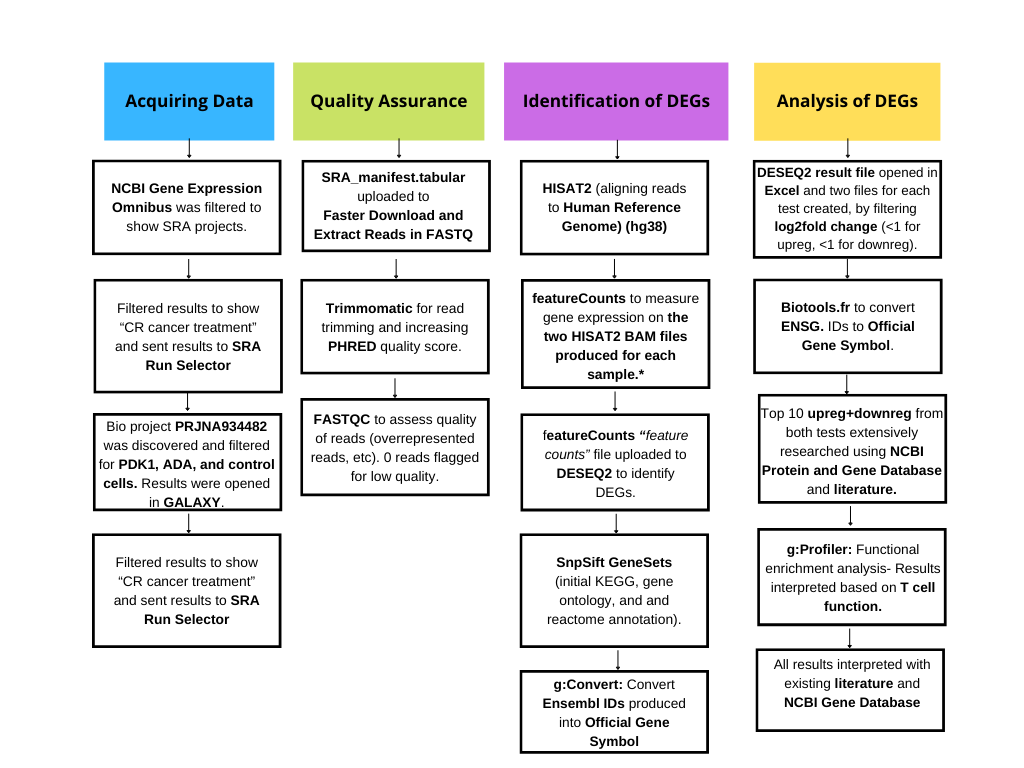
*featureCounts was utilized instead of StringTie and StringTie merge as the human genome is well annotated and does not require improvement of transcript assembly. This was for quality assurance, and I proceeded to quantify results following HISAT2.
To identify differentially expressed genes, and enriched pathways and ontologies, I utilized GALAXY's bioinformatic software and executed the pipeline above.
The data obtained for this project is single-cell RNA sequencing transcriptomes derived from public data released by Renauer et al. (2023)’s study at Yale University's Genetic Laboratory, which aimed to genetically modify T cells to combat a tumor-intrinsic evasion mechanism in which cancer cells utilize a metabolically unfavorable tumor microenvironment (TME) to suppress T cell function. Using an in silico screen, they identified Pyruvate Dehydrogenase Kinase 1 (PDK1) and Adenosine deaminase (ADA) as metabolic regulators, as gene overexpression (OE) strengthened the cytolysis of CD19-specific CD8 CAR-T cells against cognate leukemia cells, with PDK1 or ADA deficiency decreasing the cytolysis Renauer et al. (2023). Moreover, ADA-OE in α-HER2 CAR-t cells was found to increase proliferation, memory, improve cancer cytolysis, and decrease exhaustion when tested on in vivo human colorectal cancer tumors Renauer et al. (2023). In turn, ADA-OE improved tumor infiltration and clearance by α-HER2 CAR-T cells Renauer et al. (2023). By identifying the pathways and genes associated with the success of ADA-OE α-HER2 CAR-T cells in eliminating colorectal adenocarcinomas, and other solid tumors, identifying potential therapeutic targets for the treatment of colorectal adenocarcinomas and other solid tumors is possible. This project identifies differentially expressed genes and enriched pathways associated with ADA-OE α-HER2 CAR-T cells in comparison to CD19-speicifc CD8 CAR-T cells, through RNA sequencing analysis, and determines their role in the enhanced anti-colorectal cancer tumor response observed. RNA sequencing analysis was also performed on PDK1-engineered CAR-T cells, and instead the differentially expressed genes and enriched pathway's role in enhanced anti-tumor behavior were studied in the context of colorectal cancer and cognate leukemia. It was found that these CAR T-cells utilize unique metabolic pathways and replicate the mechanisms utilized by cancerous cells to allow flourishment.
Renaur et al. utilized Illumina Novaseq 6000 as their instrumental model, and purified samples following library preperation utlizing NEBNext® Poly(A) mRNA Magnetic Isolation module (Illumina) and NEBNext® Ultra™ II Directional RNA Library Prep Kit (Illumina). 3 samples of both the responding and manipulated variables were used during RNA sequencing analysis.
RNA Sequencing Analysis 1: PDK1 overexpression:
Variables:
1. Responding: CAR T-cells
2. Manipulated: PDK1 overexpressed CAR T-cells
RNA Sequencing Analysis 2: ADA overexpression:
Variables:
1. Responding: CAR T-cells
2. Manipulated: ADA overexpressed CAR T-cells
Materials:
1. National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) for transcriptomes (ncbi.nlm.nih.gov/geo/).
2. GALAXY Bioinformatics Version 24.0 (usegalaxy.org/).
3. Microsoft Excel for filteration of statistically significant differentially expressed genes.
4. Biotools.fr for conversion of ENSG IDs to Official Gene Symbol (biotools.fr/human/ensembl_symbol_converter).
5. National Center for Biotechnology Information Protein and Gene Database for interpretation of most differentially expressed genes (ncbi.nlm.nih.gov/gene).
6. g:profiler for pathway enrichment analysis and gene ontology (biit.cs.ut.ee/gprofiler/gost).
Differentially expressed genes, enriched pathways, and ontologies were considered statistically significant if their adjusted p-value = <0.05. These p-values corresponded with their Log2fold change, which measures the amount a gene is upregulated or downregulated. Upregulated genes had Log2fold changes above 0, and downregulated genes had Log2Fold changes below 0. The higher or lower this number was, the more differentially expressed the gene was. The top 10 differentially expressed genes in each group had adjusted p values = <0.0001.
A p-value calculates the probability that the results observed were due to random chance. The null hypothesis was "the investigated gene/pathway/ontology is not related to the observed difference in function of ADA/PDK1 overexpressed CAR T-cells." By adjusting the p value using the Benjamini Hochberg procedure (commonly referred to as the False Discovery Rate), the significance threshold was adjusted for each hypothesis test based on the number of tests being performed and the desired False Discovery Rate level. This ensured the results were likely correlated to the difference in CAR T-cell function, and this was supported by the literature consulted.
Consulted literature was researched in the context of colorectal cancer and T-cell function to ensure effects on global gene expression were considered.
Research
The top 10 upregulated and downregulated genes from each experiment were thoroughly analyzed and researched to uncover their role in limiting/increasing CAR T-cell success.
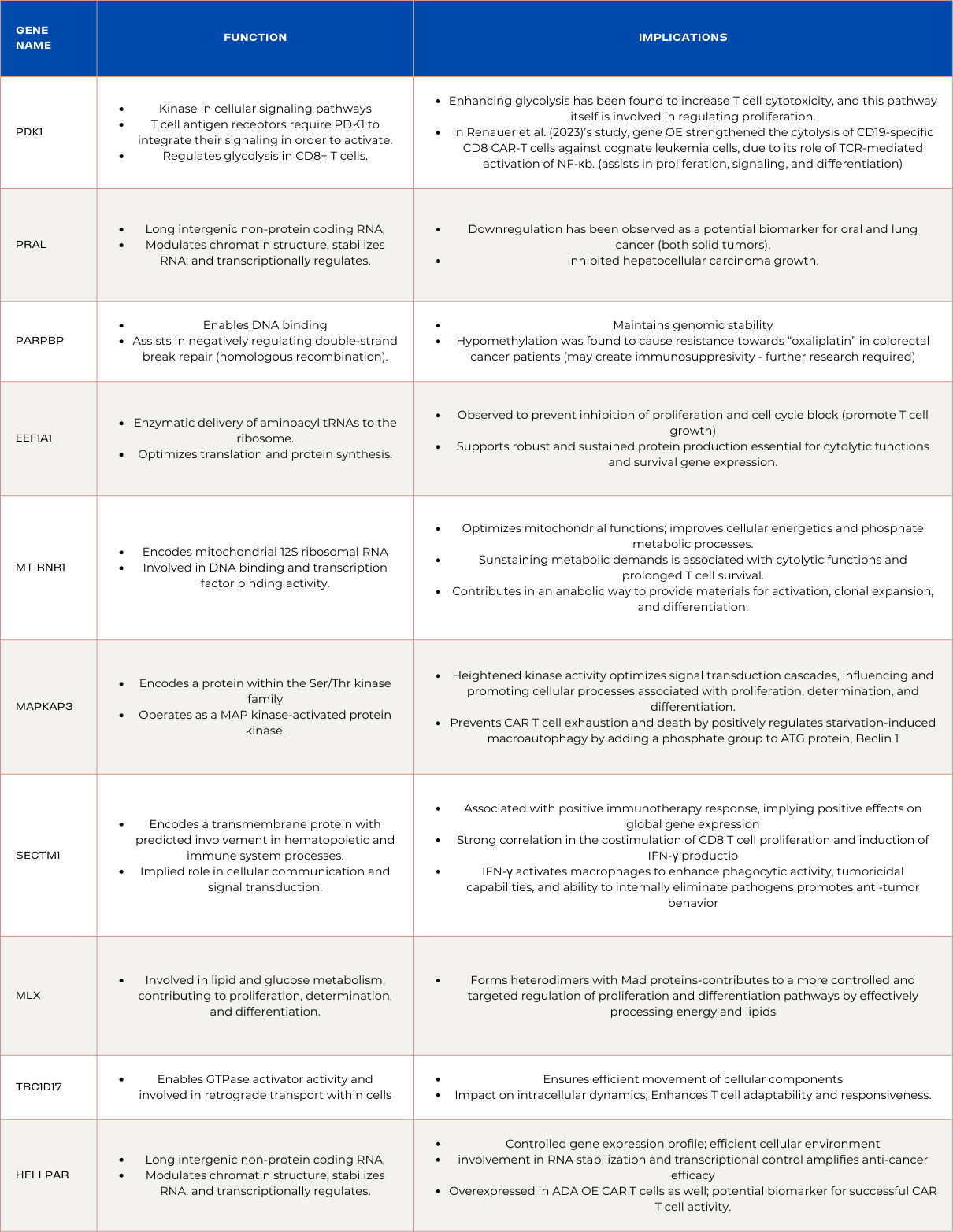
Upregulated Genes in PDK1 Overexpressed CAR T-cells
PRAL (Non-Annotated Long Intergenic Non-Protein Coding RNA):
PRAL, a non-annotated long intergenic non-protein coding RNA, participates in the modulation of chromatin structure, RNA stabilization, and transcriptional regulation. Upregulation of PRAL within PDK1-overexpressed CAR T cells implies an enhanced orchestration of these molecular processes. This heightened expression could lead to a more refined chromatin structure, stabilized RNA molecules, and precise transcriptional regulation. Downregulation of PRAL has been observed as a potential biomarker for oral and lung cancer (both solid tumors), which may be indicative of PDK1-overexpressed CAR T cells success in clearance. The optimized molecular environment created by PRAL upregulation potentially translates to improved gene expression control. Additionally, PRAL inhibited hepatocellular carcinoma growth, translating to success across solid tumors.
PARPBP (Predicted to Enable DNA Binding):
PARPBP, predicted to enable DNA binding, assists in negatively regulating double-strand break repair through homologous recombination. The upregulation of PARPBP in PDK1-overexpressed CAR T cells signifies an orchestrated enhancement in DNA binding processes, contributing to a more controlled genomic maintenance mechanism. This heightened expression potentially optimizes the negative regulation of double-strand break repair, promoting enhanced efficiency in maintaining genomic stability. The upregulation of PARPBP may foster a cellular environment with reduced error rates in DNA repair. However, its hypomethylation was found to cause resistance towards the oxaliplatin drug in colorectal cancer patients d. Further investigation is required to determine if PARPBP is involved in unfavorable prognosis of leukemia and colorectal cancer, but currently, it is not. By exerting a negative regulatory influence on repair processes, PARPBP could play a crucial role in minimizing errors and aberrations, bolstering survival gene expression and cytolytic functions in the context of colorectal cancer treatment.
EEF1A1 (Eukaryotic Elongation Factor 1 Complex Isoform A1):
EEF1A1, an isoform of the Eukaryotic Elongation Factor 1 Complex, is intricately involved in the enzymatic delivery of aminoacyl tRNAs to the ribosome. The upregulation of EEF1A1 in PDK1-overexpressed CAR T cells denotes a strategic elevation in the cellular machinery responsible for protein synthesis. This orchestrated increase in EEF1A1 expression suggests an optimization of translation processes, potentially enhancing the overall efficiency of protein synthesis within the CAR T cells. The heightened levels of EEF1A1 may contribute to an increased rate of aminoacyl tRNA delivery, supporting robust and sustained protein production essential for the cytolytic functions and survival gene expression.Although EEF1A1 is observed to prevent the inhibition of proliferation and cell cycle block in many cancers including colorectal cancer, its overexpression on the cellular level is unlikely to promote cancer growth, and the PDK1 overexpressed CAR T cells may be utilizing a successful tumor growth mechanism to promote their own proliferation. Additionally, EEF1A1 is essential in successful T cell activity, and its downregulation contributes to weakened function.
MT-RNR1 (Mitochondrially Encoded 12S RNA):
MT-RNR1, responsible for encoding mitochondrial 12S ribosomal RNA, plays a pivotal role in various cellular processes, including DNA binding and transcription factor binding activity. In the context of PDK1-overexpressed CAR T cells, the upregulation of MT-RNR1 indicates an enhancement in cellular processes associated with mitochondrial activity. The increased expression of MT-RNR1 potentially optimizes mitochondrial functions , leading to improved cellular energetics and phosphate metabolic processes. This optimization in mitochondrial activity aligns with the broader goal of PDK1-overexpressed CAR T cells, where enhanced energy production is critical for sustaining the metabolic demands associated with cytolytic functions and prolonged T cell survival. Furthermore, the upregulation of MT-RNR1 may contribute to a more controlled regulation of cellular processes. The mitochondria plays roles in lipid synthesis, calcium regulation, signaling, and cell cycle progression. In the context of T cells, they contribute in an anabolic way to provide materials for activation, clonal expansion, and differentiation of T cells.
MAPKAPK3 (MAP Kinase-Activated Protein Kinase 3):
MAPKAPK3, encoding a protein within the Ser/Thr kinase family, operates as a mitogen-activated protein kinase (MAP kinase)-activated protein kinase. In the context of PDK1-overexpressed CAR T cells, the upregulation of MAPKAPK3 suggests an orchestrated enhancement in kinase activity, contributing to effective T cell function. The heightened kinase activity may lead to optimized signal transduction cascades, influencing and promoting cellular processes associated with proliferation, determination, and differentiation.This optimized signaling aligns with the therapeutic goals of PDK1-overexpressed CAR T cells, as effective signal transduction is vital for robust T cell responses against colorectal cancer cells. Additionally, MAPKAPK3 prevents CAR T cell exhaustion and death as it, along with MAPKAPK2, positively regulates starvation-induced macroautophagy by adding a phosphate group to the crucial ATG protein, Beclin 1. With MAPKAPK3 upregulated in both ADA and PDK1 overexpressed CAR T cells, it is likely a leading reason for the increased efficiency of their function and should be investigated in its potential for CAR T cell genetic design.
SECTM1 (Transmembrane Protein in Hematopoietic and Immune System Processes):
SECTM1, encoding a transmembrane protein with predicted involvement in hematopoietic and immune system processes, demonstrates upregulation in PDK1-overexpressed CAR T cells. This upregulation suggests an orchestrated enhancement in SECTM1 expression, contributing to the modulation of immune responses. The transmembrane nature of SECTM1 implies its role in cellular communication and signal transduction. The elevated expression in PDK1-overexpressed CAR T cells indicates an optimized immune response regulation. SECTM1's broad expression in specific tissues signifies a targeted impact, potentially influencing the cellular microenvironment during the combat against colorectal cancer. Moreover, SECTM1 is associated with positive immunotherapy response, implying it has largely positive effects on global gene expression. This is due to its strong correlation in the costimulation of CD8 T cell proliferation and induction of IFN-γ production. IFN-γ primarily activates macrophages to enhance their phagocytic activity, tumoricidal capabilities, and ability to internally eliminate pathogens is vital for promoting anti-tumor behavior.
MLX (Regulator of Proliferation, Determination, and Differentiation):
MLX, a gene involved in lipid and glucose metabolism, contributing to proliferation, determination, and differentiation exhibits upregulation in PDK1-overexpressed CAR T cells. The upregulation of MLX implies an orchestrated enhancement in its regulatory role, influencing cellular processes crucial for anti-cancer activity. MLX forms heterodimers with Mad proteins, and its upregulation may contribute to a more controlled and targeted regulation of proliferation and differentiation pathways by effectively processing energy and lipids, which contribute to T cell proliferation. This heightened regulatory activity aligns with the desired therapeutic goals of PDK1-overexpressed CAR T cells. The upregulation of MLX signifies an optimized cellular response to stimuli, contributing to enhanced functionality and anti-cancer effects in PDK1-overexpressed CAR T cells.
TBC1D17 (Regulator of GTPase Activity and Retrograde Transport):
TBC1D17, a gene predicted to enable GTPase activator activity and involved in retrograde transport within cells, demonstrates upregulation in PDK1-overexpressed CAR T cells. This upregulation suggests an orchestrated enhancement in the regulation of cellular processes associated with intracellular transport and GTPase activity. The heightened GTPase activator activity may contribute to a more controlled modulation of retrograde transport, ensuring efficient movement of cellular components within PDK1-overexpressed CAR T cells. Additionally, the involvement of TBC1D17 in retrograde transport signifies a potential impact on intracellular dynamics of PDK1-overexpressed CAR T cells, enhancing their adaptability and responsiveness.
PDK1 (3-Phosphoinositide-Dependent Protein Kinase 1):
PDK1, a pivotal kinase in cellular signaling pathways, undergoes upregulation in PDK1-overexpressed CAR T cells. This upregulation plays a crucial role in enhancing T cell metabolism, cytolysis of cancer cells, and the expression of survival genes. This is showcased in the context of CD28, a corereceptor on T cells, and T cell antigen receptors require PDK1 to integrate their signaling in order to activate the T cells. When PDK1 was removed, TCR-CD28 signals could not induce NF-κB activation or protein kinase C θ phosphorylation, which are essential for T cell activation. Upregulation of PDK1 leads to an optimized metabolic state within CAR T cells and positive global gene expression as a metabolic regulator, ensuring a robust and sustained energy supply for their anti-cancer activities. The increased expression of survival genes further supports the longevity and efficacy of these CAR T cells in the challenging tumor microenvironment. Consequently, the upregulation of PDK1 in CAR T cells not only enhances their immediate cytolytic capabilities but also contributes to sustained functionality, fostering a more effective and durable response against colorectal cancer.
HELLPAR (Long Intergenic Non-Protein Coding RNA):
HELLPAR, a long intergenic non-protein coding RNA, experiences upregulation in PDK1-overexpressed CAR T cells. This upregulation assumes a pivotal role in chromatin remodeling, RNA stabilization, and transcription regulation. The heightened expression of HELLPAR signifies an orchestrated enhancement in these molecular processes, fostering a more controlled gene expression profile. This optimized chromatin structure ensures precise transcriptional regulation, contributing to a finely-tuned and efficient cellular environment within PDK1-overexpressed CAR T cells. The upregulation of HELLPAR, with its involvement in RNA stabilization and transcriptional control, potentially amplifies the anti-cancer efficacy of these CAR T cells. Moreover, HELLPAR was overexpressed in ADA overexpressed CAR T cells as well, serving as a potential biomarker for successful CAR T cell activity for leukemia and colorectal cancer.
Summary of Genes and Functions:
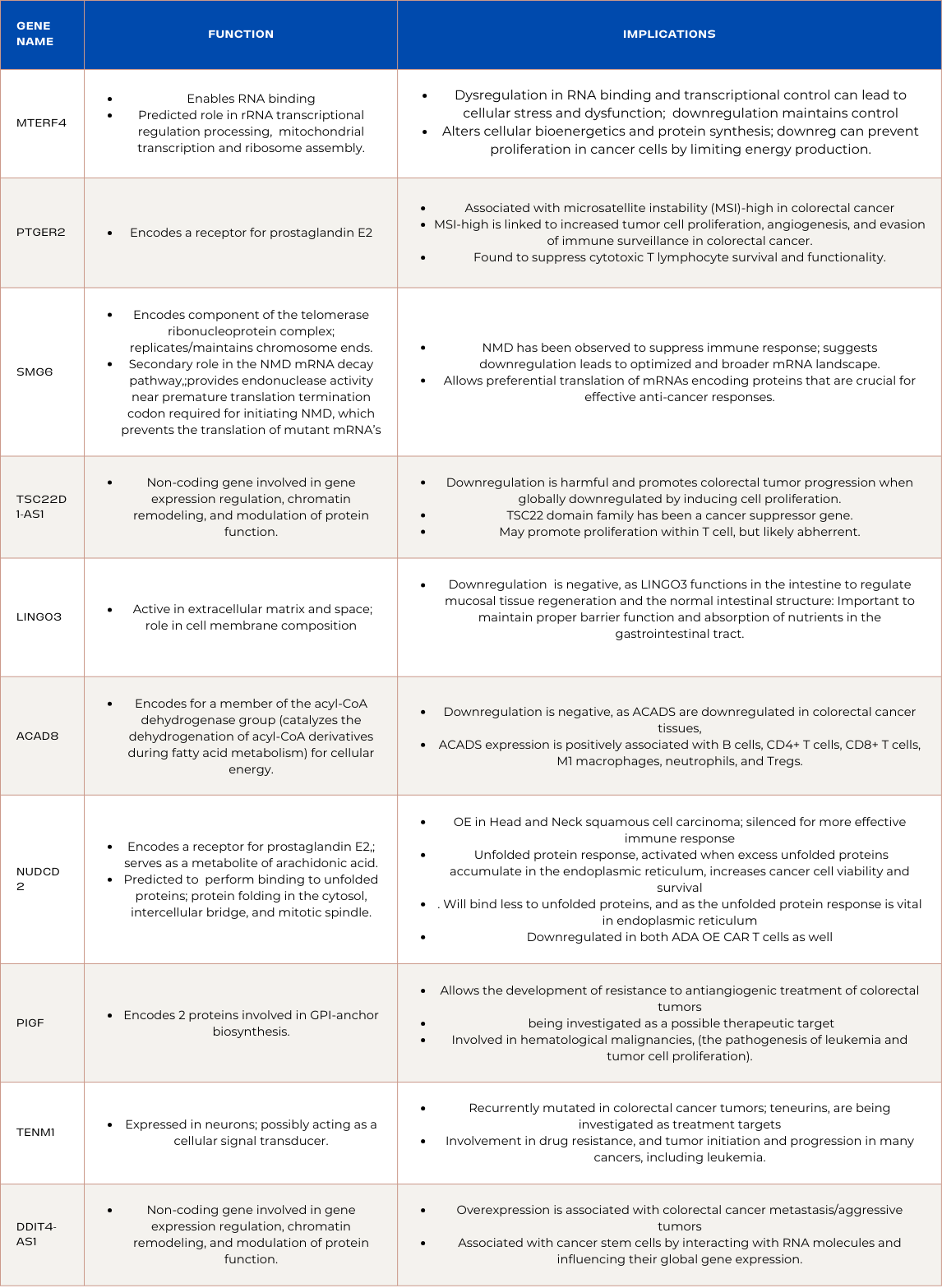
Downregulated Genes in PDK1 Overexpressed CAR T-cells
MTERF4:
Function and Implications:
MTERF4 is involved in RNA binding and plays a predicted role in rRNA transcriptional regulation and processing. Downregulation of MTERF4 in ADA-overexpressed CAR T cells suggests an optimized environment for transcriptional activity. Dysregulation in RNA binding and transcriptional control can lead to cellular stress and dysfunction. By downregulating MTERF4, ADA-overexpressed CAR T cells aim to maintain a more controlled gene expression profile, reducing the potential harmful effects associated with uncontrolled transcriptional activity. This refined transcriptional regulation could contribute to improved cellular function. However, loss of MTERF4 also leads to a decrease in mitochondrial translation, which is essential to increased mitochondrial mass and function of CD8* T cells during a time when endogenous or exogenous pyrogens elevate the body's thermoregulatory set-point. Downregulation may be indicative of damage being caused by the PDK1 overexpression, and explains why it was ineffective against colorectal cancer, as blood cancers are easier to treat.
PTGER2:
Function and Implications:
PTGER2 encodes a receptor for prostaglandin E2, and PTGER2 is associated with microsatellite instability (MSI)-high in colorectal cancer. In the context of ADA-overexpressed CAR T cells treating colorectal cancer, downregulation of PTGER2 signifies a strategic modulation of prostaglandin E2 signaling. MSI-high is linked to increased tumor cell proliferation, angiogenesis, and evasion of immune surveillance in colorectal cancer. Moreover, PTGER2 was found to suppress cytotoxic T lymphocyte survival and functionality. This is heightened when cells are affected by viruses, as it is upregulated during viral infiltration, causing the cytotoxic T cells to deteriorate. By downregulating PTGER2, ADA-overexpressed CAR T cells aim to avoid activating these pathways, enhancing their anti-cancer efficacy. The intentional disruption of prostaglandin signaling contributes to a targeted and controlled cellular response, potentially minimizing harmful effects associated with unregulated signaling in the tumor microenvironment.
SMG6:
Function and Implications:
SMG6 encodes a component of the telomerase ribonucleoprotein complex, playing a vital role in replicating and maintaining chromosome ends. In ADA-overexpressed CAR T cells designed to combat colorectal cancer, downregulation of SMG6 offers a deliberate adjustment in telomerase and chromosome maintenance. This fine-tuning aims to optimize the balance between cell proliferation and survival, contributing to an environment where ADA-overexpressed CAR T cells can efficiently target cancer cells. SMG6 has a secondary role in the nonsense-mediated mRNA decay (NMD) pathway, which provides endonuclease activity near premature translation termination codon required for initiating NMD, which prevents the translation of mutant mRNA’s with these premature termination codons. NMD has been observed to suppress immune response, which suggests its downregulation can lead to an optimized and broader mRNA landscape. This landscape could contribute to several positive outcomes, as it allows the preferential translation of mRNAs encoding proteins that are crucial for effective anti-cancer responses. Additionally, the modulation of NMD through SMG6 downregulation might lead to increased translation of specific immune-related genes, potentially enhancing the overall immune response.
TSC22D1-AS1:
Function and Implications:
TSC22D1-AS1 is a non-coding gene intricately involved in gene expression regulation, chromatin remodeling, and modulation of protein function. In the context of ADA-overexpressed CAR T cells for colorectal cancer treatment, the downregulation of TSC22D1-AS1 is harmful and promotes colorectal tumor progression when globally downregulated by inducing cell proliferation. This is consistent in other forms of cancer as the TSC22 domain family has been a cancer suppressor gene, and is likely a side effect of PDK1 overexpression in CAR T cells.
LINGO3:
Function and Implications:
LINGO3, predicted to be active in the extracellular matrix and space, plays a crucial role in cell membrane composition. Its down regulation in PDK1 overexpressed CAR T cells is negative, as LINGO3 functions in the intestine to regulate mucosal tissue regeneration and the normal intestinal structure . This is important to maintain proper barrier function and absorption of nutrients in the gastrointestinal tract.
ACAD8:
Function and Implications:
ACAD8, encodes genetic information for a member of the acyl-CoA dehydrogenase group that catalyzes the dehydrogenation of acyl-CoA derivatives during fatty acid metabolism for cellular energy. In the context of ADA-overexpressed CAR T cells combating colorectal cancer, the downregulation of ACAD8 is negative, as ACADS are downregulated in colorectal cancer tissues, and ACADS expression is positively associated with B cells, CD4+ T cells, CD8+ T cells, M1 macrophages, neutrophils, and Tregs. ACAD8, and ACAD groups in general can become a therapeutic target for CAR T cells, and are prognostic for tumor progression.
NUDCD2:
Function and Implications:
NUDCD2's main function is encoding a receptor for prostaglandin E2, which serves as a metabolite of arachidonic acid. It is also predicted to be capable of binding to unfolded proteins, and protein folding in the cytosol, intercellular bridge, and mitotic spindle. While this downregulation might suggest a modulation of protein folding processes. Although there is little information regarding NUDCD2 and prognosis in the context of colorectal cancer, it is overexpressed in Head and Neck squamous cell carcinoma, a similar solid cancer, implying it may have detrimental effects to colorectal cancer progression and thus was silenced for more effective immune response . Additionally, unfolded protein response, activated when excess unfolded proteins accumulate in the endoplasmic reticulum, increases cancer cell viability and survival during critical moments. As NUDCD2 is downregulated, it will bind less to unfolded proteins, and as the unfolded protein response is critical in the endoplasmic reticulum, its potential benefits are nullified. NUDCD2 is downregulated in both ADA overexpressed CAR T cells and PDK1 overexpressed CAR T cells, implying its suppression is crucial for effective CAR T cell activity.
PIGF:
Function and Implications:
PIGF provides the genetic information for 2 proteins involved in GPI-anchor biosynthesis, is a gene with potential implications for ADA-overexpressed CAR T cells in the context of treating colorectal cancer. PIGF was found to allow the development of resistance to antiangiogenic treatment of colorectal tumors, and is being looked at as a possible therapeutic target to enhance treatment. Moreover, PIGF has been involved in many hematological malignancies, including the pathogenesis of leukemia and tumor cell proliferation. Downregulation of PIGF suggests a modulation in GPI-anchor biosynthesis processes. This alteration may influence the composition of cellular membranes and their interactions. In the specific environment of ADA-overexpressed CAR T cells, this could contribute to a cellular milieu less conducive to colorectal cancer progression.
TENM1:
Function and Implications:
TENM1, a gene expressed in neurons and possibly acting as a cellular signal transducer, holds significance in the landscape of ADA-overexpressed CAR T cells targeting colorectal cancer. The downregulation of TENM1 could impact cellular signaling, particularly in the context of immune response and anti-cancer activity. TENM1 was recurrently mutated in colorectal cancer tumor tissue, and it’s family, the teneurins, are being investigated as potential treatment targets. Furthermore, TENM1 and its teneurin family were found to be involved in drug resistance, and tumor initiation and progression in many cancers, including leukemia.
DDIT4-AS1:
Function and Implications:
DDIT4-AS1 is a non-coding gene involved in gene expression regulation, chromatin remodeling, and modulation of protein function, and plays a crucial role in regulating gene expression within ADA-overexpressed CAR T cells. However, its overexpression has been associated with colorectal cancer metastasis, and more aggressive tumors. Additionally, it is generally associated with cancer stem cells by interacting with other RNA molecules and influencing their global gene expression.
Summary of Genes and Functions:
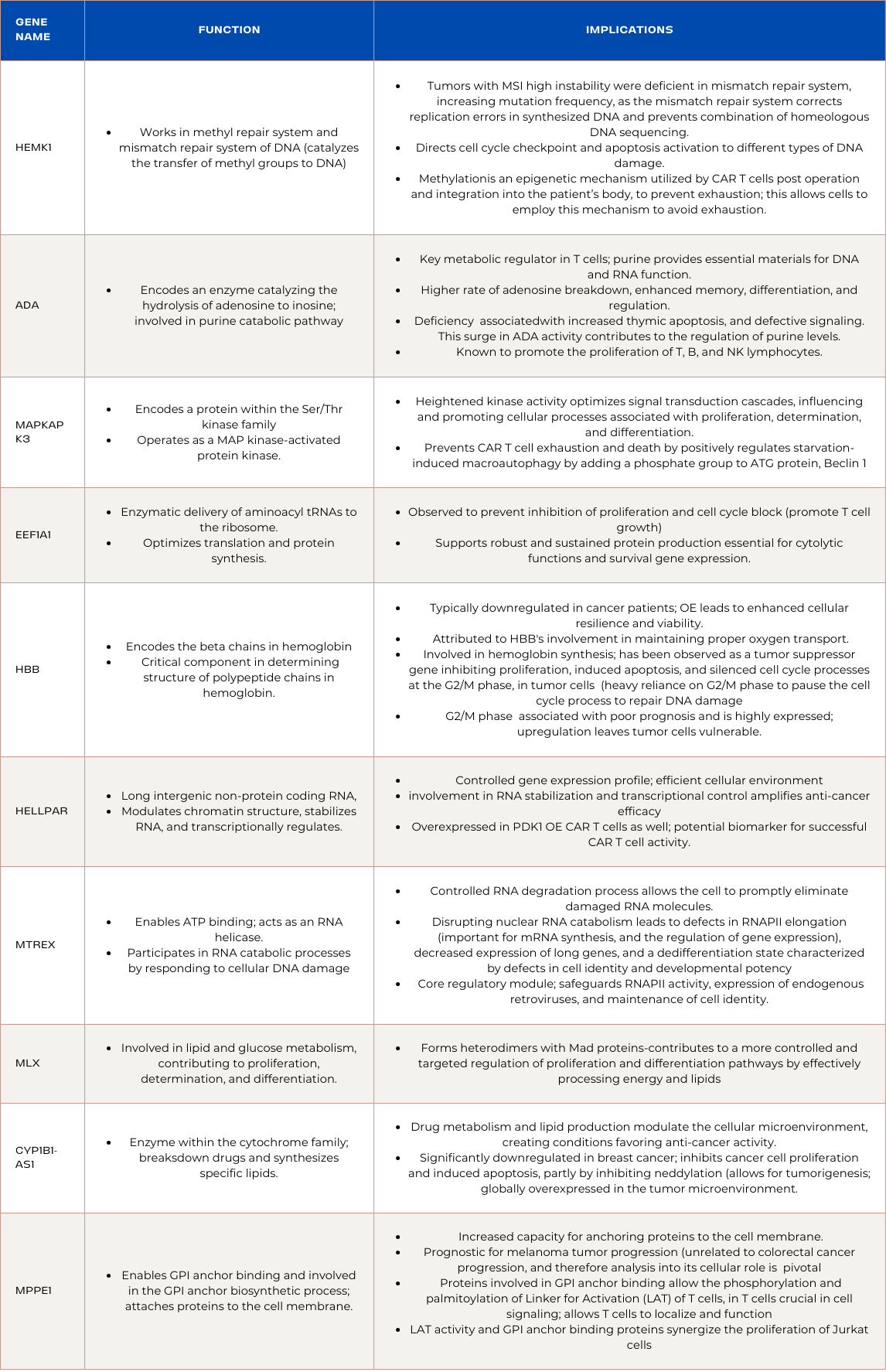
Upregulated Genes in ADA Overexpressed CAR T-cells
HEMK1:
Function and Implications:
HEMK1 works in the methyl repair system and mismatch repair system of DNA by catalyzing the transfer of methyl groups to DNA and plays a crucial role in maintaining genome stability. Tumors displaying MSI high instability had a deficiency in the mismatch repair system, creating an elevation in mutation frequency, as the mismatch repair system corrects replication errors in synthesized DNA, as well as prevents the combination of homeologous DNA sequencing. The mismatch repair system also directs cell cycle checkpoint and apoptosis activation to different types of DNA damage.This may mean that ADA overexpressed CAR T cells require this increased ability to silence and repair newly synthesized DNA, as without it, the CAR T cell is prone to avoidable exhaustion and harmful mutations. Moreover, many cancer types utilize the mechanisms HEMK1 is capable of to allow for effective metabolism, including cellular detoxification, cell and tissue growth, neurotransmitter synthesis, and gene expression. ADA overexpressed CAR T-cells potentially benefit from this methylation as the TMI’s dynamic environment can be combatted through the silencing of tumor progression genes. This may imply that ADA overexpressed CAR T cells owe their efficiency to methylation allowing them to recognize harmful sequences and thereby silence them without affecting surrounding, healthy sequences. Methylation capabilities are also important in sustaining memory in CD4 T and CD8 T cells, as well as human lymphocytes in general, allowing them to recognize and therefore destroy previously encountered pathogens, improving immune response. Unique DNA methylation patterns are also observed in terminally differentiated effector memory CD8 T cells (TEMRA) compared to other CD8 T memory cell subtypes, emphasizing the role of DNA methylation in defining distinct memory cell populations. As methylation is an epigenetic mechanism utilized by CAR T cells post operation and integration into the patient’s body, to prevent exhaustion, its upregulation allows the cells to employ this mechanism readily and avoid exhaustion.
ADA:
Function and Implications:
ADA (Adenosine Deaminase), encoding an enzyme catalyzing the hydrolysis of adenosine to inosine, plays a pivotal role in the purine catabolic pathway. In ADA-overexpressed CAR T cells, the increased activity of ADA leads to a higher rate of adenosine breakdown. This surge in ADA activity contributes to the regulation of purine levels, potentially creating an unfavorable environment for cancer cells. Additionally, ADA is known to promote the proliferation of T, B, and NK lymphocytes. The upregulation of ADA in CAR T cells reinforces the immune response, facilitating robust anti-cancer activity. The concerted action of ADA, by both regulating purine metabolism and enhancing immune cell proliferation, synergistically supports the effectiveness of CAR T cells in combating colorectal cancer.
MAPKAPK3:
Function and Implications:
MAPKAPK3, encoding a protein within the Ser/Thr kinase family, operates as a mitogen-activated protein kinase (MAP kinase)-activated protein kinase. In the context of PDK1-overexpressed CAR T cells, the upregulation of MAPKAPK3 suggests an orchestrated enhancement in kinase activity, contributing to effective T cell function. The heightened kinase activity may lead to optimized signal transduction cascades, influencing and promoting cellular processes associated with proliferation, determination, and differentiation.This optimized signaling aligns with the therapeutic goals of PDK1-overexpressed CAR T cells, as effective signal transduction is vital for robust T cell responses against colorectal cancer cells. Additionally, MAPKAPK3 prevents CAR T cell exhaustion and death as it, along with MAPKAPK2, positively regulates starvation-induced macroautophagy by adding a phosphate group to the crucial ATG protein, Beclin 1. With MAPKAPK3 upregulated in both ADA and PDK1 overexpressed CAR T cells, it is likely a leading reason for the increased efficiency of their function and should be investigated in its potential for CAR T cell genetic design.
EEF1A1:
Function and Implications:
EEF1A1, an isoform of the Eukaryotic Elongation Factor 1 Complex, is intricately involved in the enzymatic delivery of aminoacyl tRNAs to the ribosome. The upregulation of EEF1A1 in PDK1-overexpressed CAR T cells denotes a strategic elevation in the cellular machinery responsible for protein synthesis. This orchestrated increase in EEF1A1 expression suggests an optimization of translation processes, potentially enhancing the overall efficiency of protein synthesis within the CAR T cells. Moreover, heightened levels of EEF1A1 may contribute to an increased rate of aminoacyl tRNA delivery, supporting robust and sustained protein production essential for the cytolytic functions and survival gene expression. Although EEF1A1 is observed to prevent the inhibition of proliferation and cell cycle block in many cancers including colorectal cancer, its overexpression on the cellular level is unlikely to promote cancer growth, and the PDK1 overexpressed CAR T cells may be utilizing a successful tumor growth mechanism to promote their own proliferation. Additionally, EEF1A1 is essential in successful T cell activity, and its downregulation contributes to weakened function. As EEF1A1 was upregulated in both PDK1 overexpressed CAR T cells and ADA overexpressed CAR T cells, it likely is a result of the distinct overexpression, and can be attributed to their success in clearing tumors. Understanding EEF1A1’s function in the context of exhaustion, and more in depth regarding development is crucial when determining its potential for CAR T cell genetic design.
HBB:
Function and Implications:
HBB, which encodes the beta chains in hemoglobin, is a critical component in determining the structure of polypeptide chains in hemoglobin. Upregulation of HBB is often associated with promoting cell survival. In this scenario, ADA overexpression potentially assists in the increased production of HBB, leading to improved cell viability. HBB, the gene encoding beta chains in hemoglobin, is typically downregulated in cancer patients. However, its upregulation, as observed in ADA-overexpressed CAR T cells, has been linked to promoting cell survival. The increased expression of HBB may contribute to enhanced cellular resilience and viability. This could be attributed to HBB's involvement in maintaining proper oxygen transport, crucial for sustaining cellular processes. Moreover, HBB and HBA1, both involved in hemoglobin synthesis have been observed as tumor suppressor genes, as they have been observed downregulated in acute myeloid leukemia, as they inhibited proliferation, induced apoptosis, and silenced cell cycle processes at the G2/M phase, in tumor cells. Tumor cells are heavily reliant on the G2/M phase to pause the cell cycle process in order to repair DNA damage. As for colorectal cancer, upregulation of the cell division associated 5 process in general, and specifically the G2/M phase has been associated with poor prognosis and is highly expressed, meaning HBB upregulation can leave tumor cells vulnerable, and unable to perform cell integrity processes, allowing the ADA overexpressed CAR T cells to clear them efficiently.
HELLPAR:
HELLPAR, a long intergenic non-protein coding RNA, experiences upregulation in PDK1-overexpressed CAR T cells. This upregulation assumes a pivotal role in chromatin remodeling, RNA stabilization, and transcription regulation. The heightened expression of HELLPAR signifies an orchestrated enhancement in these molecular processes, fostering a more controlled gene expression profile. This optimized chromatin structure ensures precise transcriptional regulation, contributing to a finely-tuned and efficient cellular environment within PDK1-overexpressed CAR T cells. The upregulation of HELLPAR, with its involvement in RNA stabilization and transcriptional control, potentially amplifies the anti-cancer efficacy of these CAR T cells. Moreover, HELLPAR was overexpressed in ADA overexpressed CAR T cells as well, serving as a potential biomarker for successful CAR T cell activity for leukemia and colorectal cancer.
MTREX:
MTREX, enables ATP binding and acts as an RNA helicase, participating in RNA catabolic processes by responding to cellular DNA damage, and is localized in the nucleoplasm. In ADA-overexpressed CAR T cells designed for combatting colorectal cancer, the upregulation of MTREX suggests a regulation of RNA turnover. The enzymatic activity of MTREX, as an RNA helicase, contributes to the unwinding of RNA structures, facilitating their degradation. This controlled RNA degradation process is beneficial as it allows the cell to promptly eliminate unwanted or damaged RNA molecules. Additionally, disrupting nuclear RNA catabolism leads to defects in RNAPII elongation, decreased expression of long genes, and a dedifferentiation state characterized by defects in cell identity and developmental potency. RNAPII elongation is important for mRNA synthesis, and the regulation of gene expression. This means MTREX’s role in RNA catabolism appears to be a core regulatory module that safeguards important cellular processes, including RNAPII activity, expression of endogenous retroviruses (ERVs), and maintenance of cell identity during embryonic development.
MLX: (Regulator of Proliferation, Determination, and Differentiation):
MLX, a gene involved in coding a member of the helix-loop-helix leucine zipper (bHLH-Zip) transcription factor family that is involved in lipid and glucose metabolism. These proteins also assist in proliferation, determination, and differentiation, exhibits upregulation in PDK1-overexpressed CAR T cells. The upregulation of MLX implies an orchestrated enhancement in its regulatory role, influencing cellular processes crucial for anti-cancer activity. MLX forms heterodimers with Mad proteins, and its upregulation may contribute to a more controlled and targeted regulation of proliferation and differentiation pathways by effectively processing energy and lipids, which contribute to T cell proliferation. This heightened regulatory activity aligns with the desired therapeutic goals of PDK1-overexpressed CAR T cells. The upregulation of MLX signifies an optimized cellular response to stimuli, contributing to enhanced functionality and anti-cancer effects in PDK1-overexpressed CAR T cells.
CYP1B1-AS1, an enzyme within the cytochrome family, plays a crucial role in the breakdown of drugs and the synthesis of specific lipids. Upregulation of CYP1B1-AS1 in ADA-overexpressed CAR T cells may influence drug metabolism and lipid production. This could potentially contribute to the modulation of the cellular microenvironment, creating conditions that favor the anti-cancer activity of ADA-overexpressed CAR T cells. It was observed that CYP1B1-AS1 was significantly downregulated in breast cancer, and inhibited cancer cell proliferation and induced apoptosis, partly by inhibiting neddylation. Neddylation modulates many essential biological processes, allowing for tumorigenesis, and is globally overexpressed in the tumor microenvironment.
MPPE1, predicted to enable GPI anchor binding and involved in the GPI anchor biosynthetic process, plays a critical role in attaching specific proteins to the cell membrane. Upregulation of MPPE1 in ADA-overexpressed CAR T cells suggests an increased capacity for anchoring proteins to the cell membrane. This heightened activity may influence the overall composition of the cell membrane and contribute to alterations in cellular signaling and interactions. Although MPPE1 has been found prognostic for melanoma tumor progression, it is unrelated to colorectal cancer progression, and therefore analysis into its cellular role becomes pivotal. Proteins involved in GPI anchor binding were found to allow the phosphorylation and palmitoylation of Linker for Activation (LAT) of T cells, in T cell. This protein is crucial in cell signaling, and it requires phosphorylation and palmitoylation to localize and function. Moreover, LAT activity and GPI anchor binding proteins were found to synergize the proliferation of Jurkat cells, an immortalized line of T cell lymphocytes.
Summary of Genes and Functions:
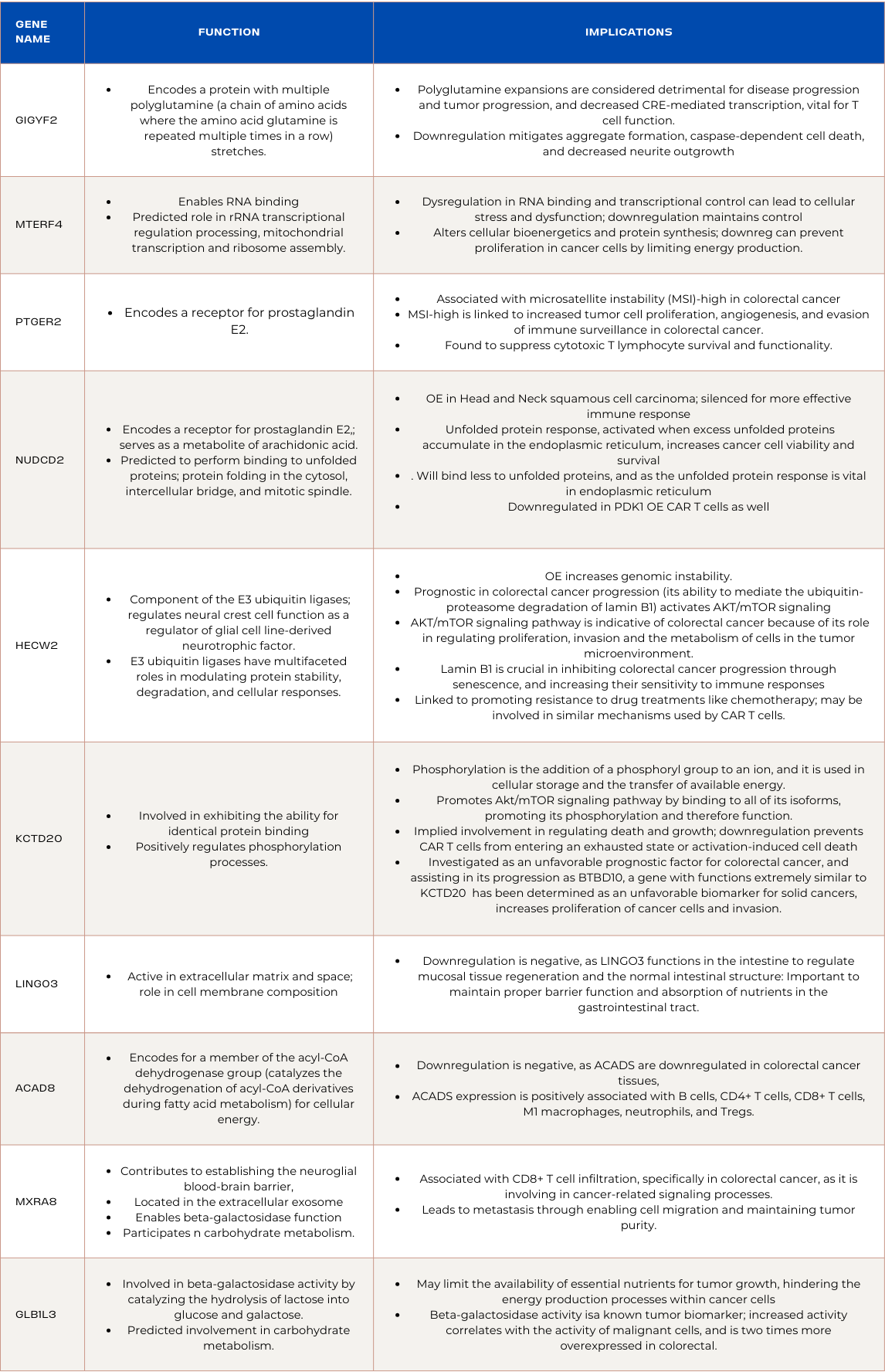
Downregulated Genes in ADA Overexpressed CAR T-cells
GIGYF2:
GIGYF2, encoding a protein with multiple polyglutamine (a chain of amino acids where the amino acid glutamine is repeated multiple times in a row) stretches, plays a role in cellular processes influenced by polyglutamine-containing proteins. Polyglutamine expansions are linked to alterations in the cellular environment, impacting cellular processes such as signal transduction, protein folding, and GIGYF2 is involved in regulating these processes.
This modulation may lead to nuanced changes in the cellular landscape, potentially affecting the dynamics of signaling cascades or protein-protein interactions. Polyglutamine expansions are considered detrimental for disease progression and tumor progression, and decreased CRE-mediated transcription, which is vital for gene expression. By downregulating GIGYF2, a protein involved in these processes, negative effects such as aggregate formation, caspase-dependent cell death, and decreased neurite outgrowth can be mitigated. By strategically influencing polyglutamine-associated cellular processes, GIGYF2 may assist in creating an environment that hinders colorectal cancer progression, but it remains non prognostic in most cancers, and little research is related to its role it T cells and the tumor microenvironment.
MTERF4:
MTERF4, a gene allowing RNA binding, is predicted to have roles in rRNA transcriptional regulation and processing, as well as mitochondrial transcription and ribosome assembly. Dysregulation in RNA binding and transcriptional control can lead to cellular stress and dysfunction. By downregulating MTERF4, ADA-overexpressed CAR T cells aim to maintain a more controlled gene expression profile, reducing the potential harmful effects associated with uncontrolled transcriptional activity. The downregulation of MTERF4 in ADA-overexpressed CAR T cells indicates a targeted impact on RNA-related processes, potentially altering cellular bioenergetics and protein synthesis. As MTERF4 is involved in the assembly of mitochondrial ribosomes, its downregulation is contextually beneficial, as it can prevent proliferation in cancer cells by limiting energy production.
PTGER2:
PTGER2 encodes a receptor for prostaglandin E2, and PTGER2 is associated with microsatellite instability (MSI)-high in colorectal cancer. In the context of ADA-overexpressed CAR T cells treating colorectal cancer, downregulation of PTGER2 signifies a strategic modulation of prostaglandin E2 signaling. MSI-high is linked to increased tumor cell proliferation, angiogenesis, and evasion of immune surveillance in colorectal cancer. Moreover, PTGER2 was found to suppress cytotoxic T lymphocyte survival and functionality. This is heightened when cells are affected by viruses, as it is upregulated during viral infiltration, causing the cytotoxic T cells to deteriorate. By downregulating PTGER2, ADA-overexpressed CAR T cells aim to avoid activating these pathways, enhancing their anti-cancer efficacy. The intentional disruption of prostaglandin signaling contributes to a targeted and controlled cellular response, potentially minimizing harmful effects associated with unregulated signaling in the tumor microenvironment.
NUDCD2:
NUDCD2's main function is encoding a receptor for prostaglandin E2, which serves as a metabolite of arachidonic acid. It is also predicted to be capable of binding to unfolded proteins, and protein folding in the cytosol, intercellular bridge, and mitotic spindle. While this downregulation might suggest a modulation of protein folding processes. Although there is little information regarding NUDCD2 and prognosis in the context of colorectal cancer, it is overexpressed in Head and Neck squamous cell carcinoma, a similar solid cancer, implying it may have detrimental effects to colorectal cancer progression and thus was silenced for more effective immune response. Additionally, unfolded protein response, activated when excess unfolded proteins accumulate in the endoplasmic reticulum, increases cancer cell viability and survival during critical moments. As NUDCD2 is downregulated, it will bind less to unfolded proteins, and as the unfolded protein response is critical in the endoplasmic reticulum, its potential benefits are nullified.
HECW2:
HECW2, an integral component of the E3 ubiquitin ligases, intricately regulates neural crest cell function as a regulator of glial cell line-derived neurotrophic factor. E3 ubiquitin ligases, including HECW2, are recognized for their multifaceted roles in modulating protein stability, degradation, and cellular responses, but overexpression has been found to increase genomic instability, which is negative. HECW2 has been found to be prognostic in colorectal cancer progression, because of its ability to mediate the ubiquitin-proteasome degradation of lamin B1, which activates AKT/mTOR signaling. The AKT/mTOR signaling pathway has been an indicator of colorectal cancer because of its role in regulating proliferation, invasion and the metabolism of cells in the tumor microenvironment. On the other hand, lamin B1 is crucial in inhibiting colorectal cancer progression through senescence, and increasing their sensitivity to immune responses. This means the downregulation of HECW2 may be linked to the effectiveness of ADA overexpressed CAR T cells. Moreover, HECW2 has been linked to promoting resistance to drug treatments like chemotherapy, which may be involved in similar mechanisms used by CAR T cells.
KCTD20:
KCTD20 is involved in exhibiting the ability for identical protein binding and the positive regulation of phosphorylation processes. Phosphorylation is the addition of a phosphoryl group to an ion, and it is used in cellular storage and the transfer of available energy. KCTD20 is also involved in promoting the Akt/mTOR signaling pathway by binding to all of its isoforms, promoting its phosphorylation and therefore function. This implies that KCTD20 may be involved in regulating death and growth, and its downregulation is to prevent the CAR T cells from entering an exhausted state or activation-induced cell death. Additionally, KCTD20 is being investigated as an unfavorable prognostic factor for colorectal cancer, and assisting in its progression. BTBD10, a gene with functions extremely similar to KCTD20 that has been determined as an unfavorable biomarker for most solid cancers, increases proliferation of cancer cells and invasion, which may be similar to how KCTD20 increases colorectal cancer progression.
LINGO3:
LINGO3 is predicted to be active in the extracellular matrix and space, and plays a crucial role in cell membrane composition. ADA overexpression influencing the downregulation of LINGO3 might impact the structural integrity of the extracellular matrix and cellular membranes. This modulation can potentially alter cellular interactions, affecting processes like adhesion and communication. Considering LINGO3's involvement in the cell membrane, its downregulation may influence cell surface interactions and signaling. Although there is limited information regarding LINGO3’s involvement in colorectal cancer, its function in composing cell membrane may be the reason for its suppression, as “membrane therapy” has emerged to suppress cancer growth factors by regulating signaling and regulate transport of materials in and out of the cells. Constant composition may disrupt this process, and suppression may regulate aberrant cell construction found in colorectal cancer.
ACAD8:
ACAD8, encodes genetic information for a member of the acyl-CoA dehydrogenase group that catalyzes the dehydrogenation of acyl-CoA derivatives during fatty acid metabolism for cellular energy. In the context of ADA-overexpressed CAR T cells combating colorectal cancer, the downregulation of ACAD8 is negative, as ACADS are downregulated in colorectal cancer tissues, and ACADS expression is positively associated with B cells, CD4+ T cells, CD8+ T cells, M1 macrophages, neutrophils, and Tregs. ACAD8 is also favorably prognostic in many solid tumors, including colorectal tumors.
MXRA8:
MXRA8 is predicted to contribute to establishing the neuroglial blood-brain barrier, located in the extracellular exosome, enabling beta-galactosidase function and participating in carbohydrate metabolism. The downregulation of MXRA8 suggests a potential modulation of immune-related processes and interactions with the tumor microenvironment. The expression of MXRA8 is associated with CD8+ T cell infiltration, specifically in colorectal cancer, as it is involving in cancer-related signaling processes. Furthermore, it led to metastasis through enabling cell migration and maintaining tumor purity. The downregulation implies that ADA overexpressed CAR T cells successfully suppressed MXRA8’s function, and may be partly how they were able to clear colorectal tumors and limit progression.
GLB1L3:
GLB1L3 is predicted to involve beta-galactosidase activity by catalyzing the hydrolysis of lactose into glucose and galactose, and is also predicted to be involved in carbohydrate metabolism. This involvement in carbohydrate metabolism suggests that GLB1L3 downregulation may limit the availability of essential nutrients for tumor growth, potentially hindering the energy production processes within cancer cells. Moreover, beta-galactosidase activity is known as a tumor biomarker, as its increased activity correlates with the activity of malignant cells, and is two times more overexpressed in colorectal cancer.
As hypothesized, differentially expressed genes in both cell types were contributing to successful CAR T cell function, and in PDK1 OE CAR T-cells case, played a role in its limitations, as well. In ADA OE CAR T-cells, functions such as the mismatch repair system to correct transcriptional errors, tumor suppressor genes inducing apoptosis; functional oxygen transport; promotion of proliferation of immune cells; controlled gene expression; RNA catabolism; metabolizers and essential synthesizers of lipids and hemoglobin; inhibition of neddylation to promote anti cancer tumor microenvironment, and GPI anchor binding to ensure the phosphorylation and palmitoylation of Linker for Activation of T cells were observed. These functions were typically associated with positive prognosis. Functions promoting dysregulation in RNA binding, MSI-high, cancer proliferation through binding to unfolded proteins, AKT/mTOR signaling pathway, and CD8+ T cell infiltration were downregulated. As for PDK1 OE CAR T-cells, genes involved in metabolic regulating processes, prognostic factors for colorectal cancer, proliferation, cytolytic functions, kinase activity for signaling transduction and differentiation, starvation induced macroautophagy, efficient cellular movement, RNA stabilization, and positive immunotherapy response were upregulated. However, PDK1 OE CAR T-cells suppressed a few genes unfavorably, which may explain its limitations in clearing colorectal cancer adenocarcinoma. Genes experiencing downregulation in PDK1 OE CAR T-cells had functions relating to limiting cancer cell energy production, MSI-high colorectal cancer, NMD mRNA decay, regulation of mucosal tissue regeneration (also downregulated in ADA OE CAR T-cells), immune cell proliferation, cancer proliferation through binding to unfolded proteins, resistance to anti-angiogenic treatment, mutation prone genes, colorectal metastasis, and noncoding RNA where downregulation is a negative prognosis factor.
Summary of Genes and Functions:
41.8% of upregulated genes found were common with the other experiment's (ADA OE or PDK1 OE) upregulated genes. Further, 35.4% of downregulated genes were common. This demonstrates the genes and pathways identified are likely contributing heavily to success, and genes that do not overlap with ADA OE are likely what are causing limitations with PDK1 OE CAR T-cells.
Pathways Enriched in ADA Overexpressed CAR T-cells:
Pathways Enriched in PDK1 Overexpressed CAR T-cells:
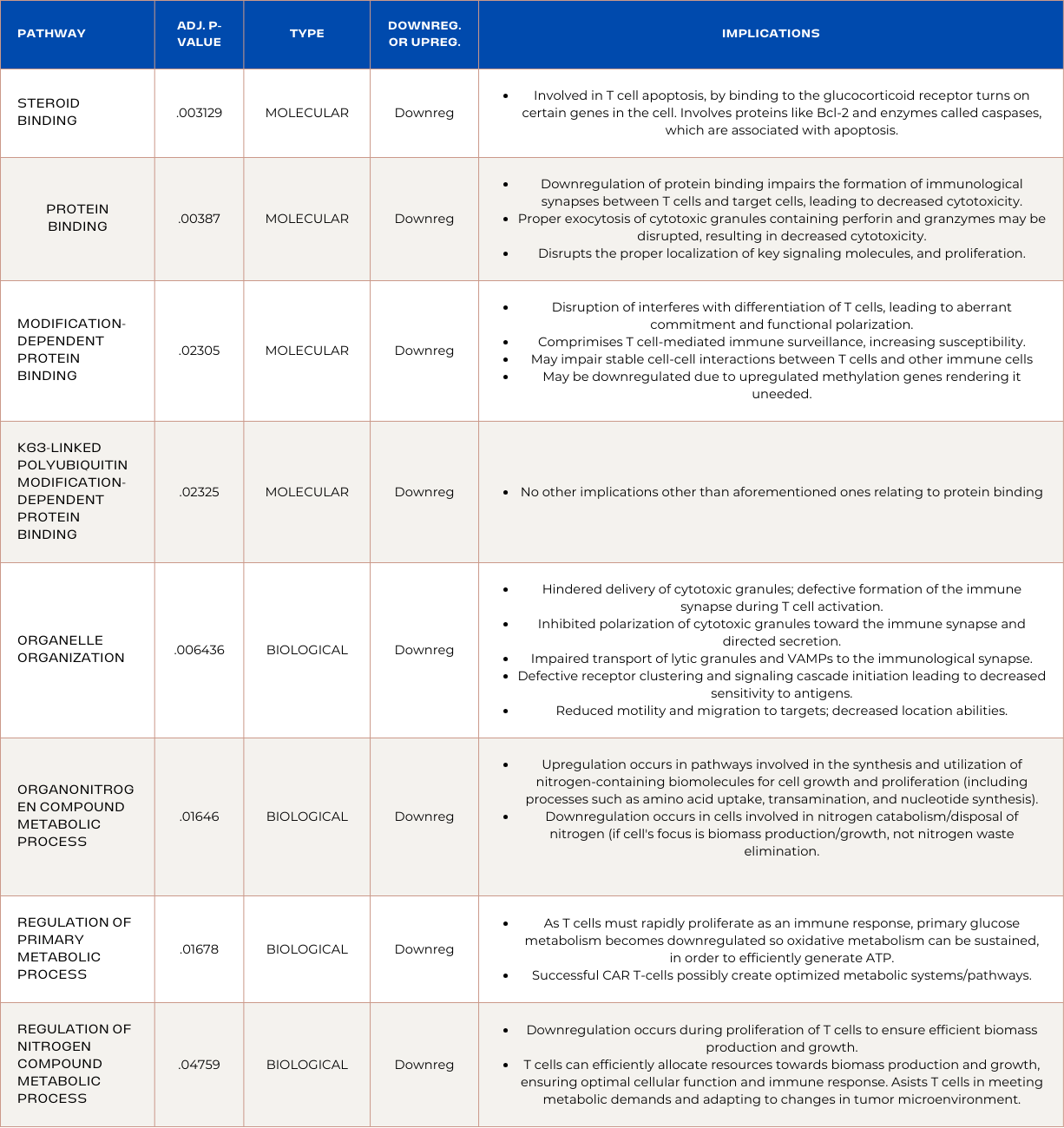
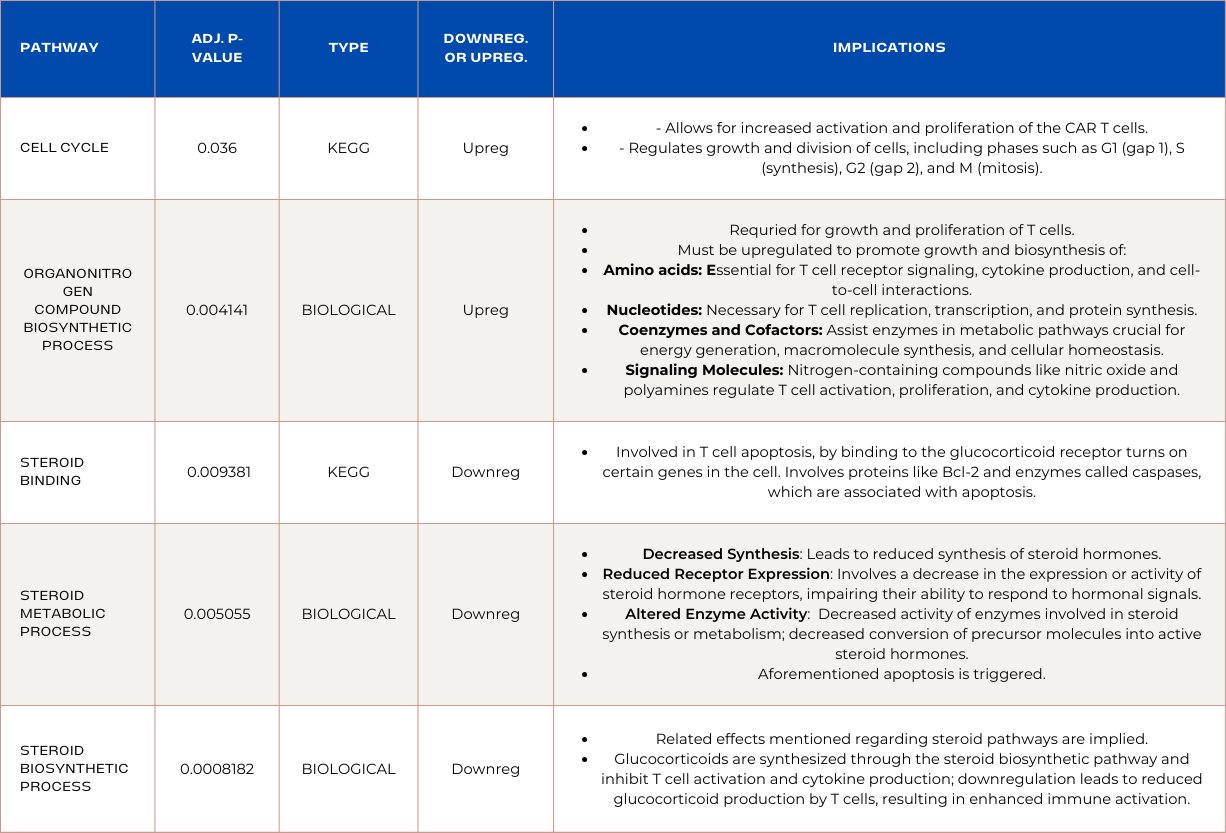
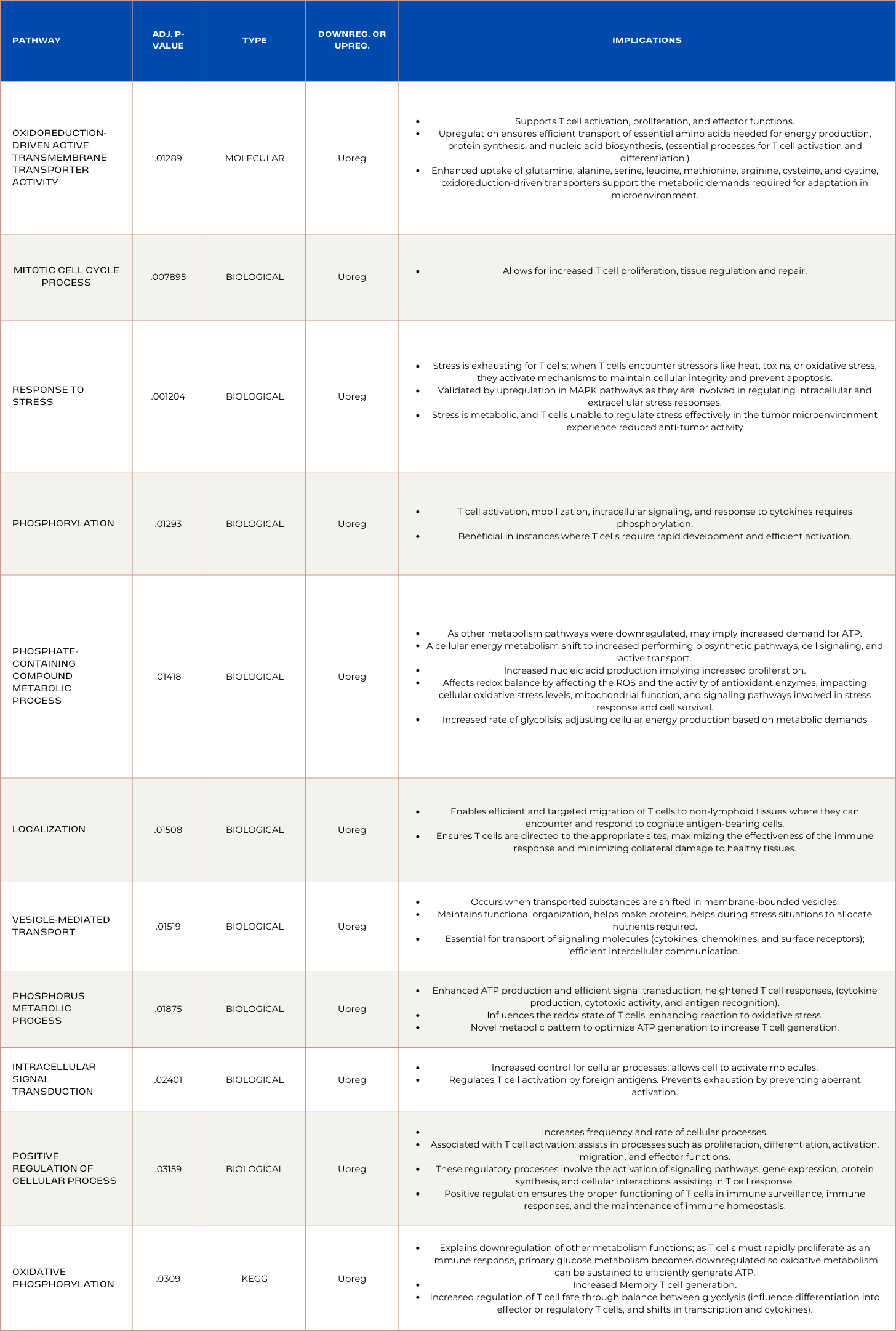
Pathway enrichment analysis revealed that RNA sequencing analysis identified a novel discovery: Successful CAR T-cells develop unique metabolic mechanisms to prioritize ATP production and limit resources used to establish multiple metabolic pathways. Phosphorous-related-metabolism pathways were prioritized due to their role in efficient signal transduction; heightened T cell responses; cytokine production; cytotoxic activity; and antigen recognition. Moreover, pathways enriched in upregulated genes prevented abhorrent activation to prevent exhaustion and enhanced signaling and cellular movement mechanisms. The upregulation of stress response is supported by the upregulation of MAPK pathways involved in intracellular and extracellular stress responses. Found in ADA OE CAR T-cells, implying their ability to maintain prolonged cytotoxicity may be attributed to stress response. It was also discovered that successful CAR T-cells exploit tumor mechanisms like upregulated cell cycle pathway, and mitotic cell cycle process. Lastly, down regulation of steroid related pathways inhibition are novel indicators of CAR T-cell persistence and are involved in apoptosis as they lead to reduced glucocorticoid production by T cells, resulting in enhanced immune activation. These discoveries are novel and are specific to colorectal cancer clearance, which can transform the way we approach CAR T-cell treatment for solid tumors.
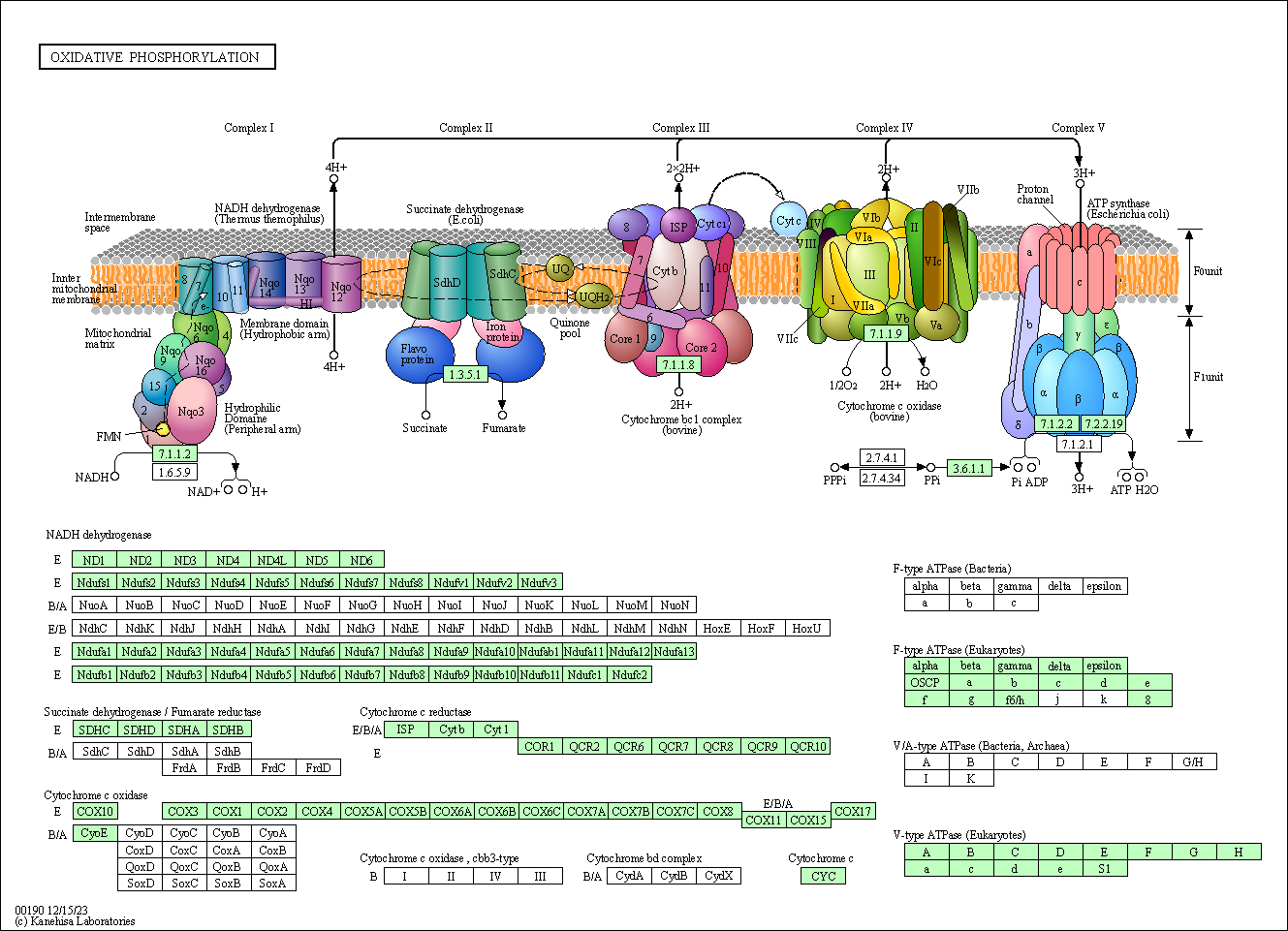
Comprehensive visuals of the interconnected genes involved in the enrichment of a pathway were generated by DAVID, and can allow for specified designing that modulates genes with adverse effects on global gene landscape. Below is a DAVID generated visual of the pathway "oxidative phosphorylation". It depicts the most significantly modulated genes, as well as the genes will larger roles in the pathway. This is crucial for in silico screening and was achieved through this project.
Data
The data utilized is around 5 gigabytes per sample. Therefore, I will provide screenshots to serve as an example of what the data looked like during each stage of analysis. Data was obtained from the NCBI GEO Bioproject "PRJNA934482". Under this Bioproject, the following samples were analyzed:
Control Sample Names:
1. GSM7040938
2. GSM7040937
3. GSM7040936
PDK1 Overexpressed Sample Names:
1. GSM7040941
2. GSM7040940
3. GSM7040939
ADA Overexpressed Sample Names:
1. GSM7040935
2. GSM7040934
3. GSM7040933
First Page of Raw Data:

First Page of Data Following Trimmomatic Operation:

(As reads were high quality, the data remained unchanged as PHRED scores were extremely high.)
First Page of FAST QC (Quality Assurance) Reports:
First Page of BAM Files (HISAT2 Alignment):
First Page of the featureCounts "Counts" File:
First Page of the DESEQ2 Results File:
Example of DESEQ2 Result File Filtering to Represent Only Statistically Significant Differentially Expressed Genes:
The rest of the data was processed in the form of ENSG IDs and official gene symbols.
Conclusion
CONCLUSION
With colorectal cancer incidence continually rising, developing effective CAR T-cell therapy for solid tumor clearance is imperative. The identification of differentially expressed genes, and commonalities between ADA OE CAR T-cells and PDK1 OE CAR T-cells in affecting global gene expression has implications ranging from biomarker identification, to indications of CAR T-cell persistence and exhaustion. Moreover, differential gene expression revealed limitations in clearing solid tumors, including silencing of genomic stabilizing genes and inhibition of mucosal tissue regulators. Indicators for positive CAR T-cell activity were linked to the downregulation of apoptosis-inducing pathways, nonsense-mediated mRNA decay, neddylation, and proliferation inhibiting genes, as well as the upregulation of pathways involved in rapid cellular cycle processes, genomic stability regulators, efficient metabolizers, increased cytokine production/increased cytolytic functions, and protection against CD8+ T cell infiltration. This study discovered that successful CAR T-cells exploit tumor mechanisms like upregulated cell cycle pathways, mitotic cell cycle processes, and heightened stress responses as well as preventing against abhorrent activation and enhanced signaling and cellular movement to conserve energy. A novel discovery derived from pathway analysis was the enrichment of phosphorous related metabolism pathways and downregulation of other metabolic processes including steroid and nitrogen related. This is hypothesized to prioritize efficient signal transduction, heightened T cell responses, cytokine production, cytotoxic activity, and antigen recognition. These results are imperative as they identify genes affecting global gene expression most drastically and identifies novel therapeutic targets and mechanisms differing from blood-cancer specific CAR T-cells which can assist in developing FDA approved solid tumor treatment and assessing theorized patient response based on identified biomarkers.
FUTURE DIRECTIONS
Although the analytical methods in this study were quality assured through the avoidance of tools like “StringTie” to ensure proper quantification, and p-value was adjusted using the Benjimini Hochberg method, this approach has limitations.
- Limited information regarding effects of genes on solid tumor-specific CAR T-cells: As this field is emerging, literature was limited as biomarkers and contributors were novel and were mostly researched related to function rather than the official gene symbol. This may mean that without in silico validation, conclusions derived are limited in specificity.
- Sample size and replication: Renauer et al. (2023)’s study specifies they utilized 3 biological replicates per cell but does not specify sample size (only stating it is in the scope of similar studies). Understanding variation in patient underlying disease and various pre-existing genetic traits is crucial for validation of results.
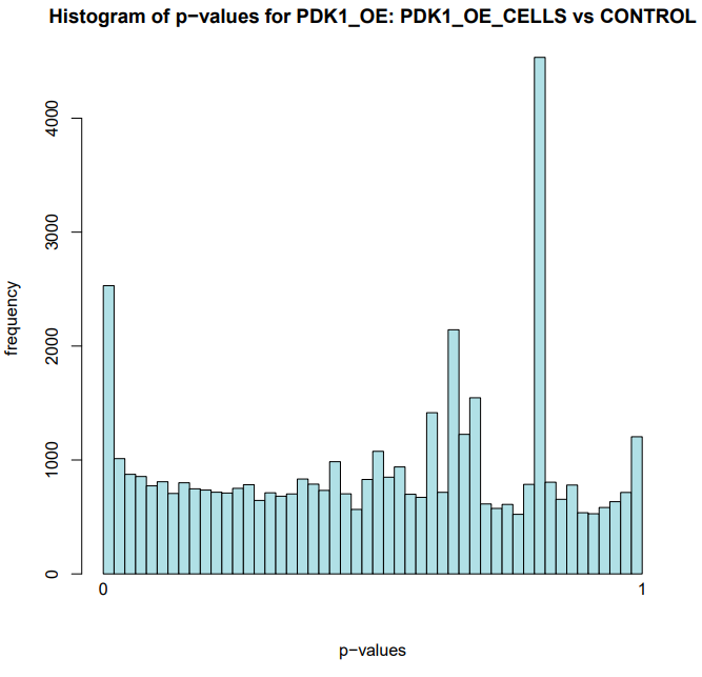
Histogram plot of p-values for PDK1 OE CAR T-cells reveals methodology excluded numerous false positives through extensive quality assurance (trimming of lower quality reads, adjusted p-value calculation).
In silico Validation
In silico screens are utilized to simulate biological experiments and identify the behavior of the subject when interacting with a certain compound.
Employ experimental validation techniques such as quantitative real-time polymerase chain reaction (qRT-PCR), Western blotting, and functional assays (e.g., proliferation assays, apoptosis assays) to validate the functional significance of specific genes and pathways in CAR T cell therapy.
- Integrate multiple layers of omics data, including proteomic and genomic data
- Apply machine learning algorithms such as random forest, support vector machines, or neural networks to prioritize candidate biomarkers based on their predictive power and discriminative ability.
- Explore drug-gene interaction databases and pharmacogenomic resources to identify novel therapeutic targets that may modulate key biological processes.
- Additional criteria such as druggability, specificity, and safety profiles could be considered when prioritizing therapeutic targets for further experimental validation and clinical investigation.
Differentially Expressed Gene's Role(s) in Global Gene Expression Using Metabolomics
Metabolic Pathway Crosstalk:
- Identify alterations in metabolic pathways associated with DEGs from RNA-seq analysis.
- Explore the correlation between gene expression changes and metabolic adaptations.
Identification of Metabolic Signatures:
- Correlate changes in metabolite abundance with DEG expression levels.
- Pinpoint metabolites indicative of gene dysregulation and downstream metabolic effects.
Validation of Transcriptomic Findings:
- Use metabolomic data as independent validation for RNA-seq analysis results.
- Confirm consistent changes in metabolite profiles corresponding to DEGs.
Prediction of Gene Function and Pathway Activity:
- Inform predictions of gene function and pathway activity based on metabolic phenotypes associated with DEGs.
- Prioritize candidate genes and pathways for functional and in silico validation experiments.
What’s Next?
To further understand the mechanisms involved in successful CAR T-cell clearance of colorectal cancer and other solid tumors, experimenting with biomarkers identified and leaders in enriched pathways using in silico screens can advance our understanding on specific DEGs role in gene expression and mechanisms, which may lead to the identification of genes catalyzing CAR T-cell persistence more than ADA and PDK1. Moreover, limitations of PDK1 identified should be used to inform clinical trials ran, by avoiding triggering identified novel pathways. Lastly, bulk transcriptomic analysis of colorectal tumor the CAR T-cells were tested on can be performed to identify signatures in tumor microenvironment leading to reduced infiltration and vulnerability to T cells, as well as effects of altered CAR T-cells on overall tumor gene expression. RNA sequencing analysis can also be performed on patients receiving ADA/PDK1 overexpressed CAR T-cells to identify biomarkers associated with remission and relapse.
Citations
-
Sterner, R. C., & Sterner, R. M. (2021, April 6). Car-T cell therapy: Current limitations and potential strategies. Nature News. https://www.nature.com/articles/s41408-021-00459-7
-
Johnson, A., Townsend, M., & O'Neill, K. (2022). Tumor Microenvironment Immunosuppression: A Roadblock to CAR T-Cell Advancement in Solid Tumors. Cells, 11(22), 3626. https://doi.org/10.3390/cells11223626
-
World Health Organization. (2023). Colorectal Cancer (11th ed.). https://www.who.int/
-
O'Sullivan, D. E., Hilsden, R. J., Ruan, Y., Forbes, N., Heitman, S. J., & Brenner, D. R. (2020). The incidence of young-onset colorectal cancer in Canada continues to increase. Cancer epidemiology, 69, 101828. https://doi.org/10.1016/j.canep.2020.101828
-
Renauer, P., Park, J. J., Bai, M., Acosta, A., Lee, W. H., Lin, G. H., Zhang, Y., Dai, X., Wang, G., Errami, Y., Wu, T., Clark, P., Ye, L., Yang, Q., & Chen, S. (2023). Immunogenetic Metabolomics Reveals Key Enzymes That Modulate CAR T-cell Metabolism and Function. Cancer immunology research, 11(8), 1068–1084. https://doi.org/10.1158/2326-6066.CIR-22-0565
-
Sayers, E. W., Bolton, E. E., Brister, J. R., Canese, K., Chan, J., Comeau, D. C., Connor, R., Funk, K., Kelly, C., Kim, S., Madej, T., Marchler-Bauer, A., Lanczycki, C., Lathrop, S., Lu, Z., Thibaud-Nissen, F., Murphy, T., Phan, L., Skripchenko, Y., Tse, T., … Sherry, S. T. (2022). Database resources of the national center for biotechnology information. Nucleic acids research, 50(D1), D20–D26. https://doi.org/10.1093/nar/gkab1112
-
Huang, Z., Zhou, J. K., Peng, Y., He, W., & Huang, C. (2020). The role of long noncoding RNAs in hepatocellular carcinoma. Molecular cancer, 19(1), 77 . https://doi.org/10.1186/s12943-020-01188-4
-
Hong, B., Lu, R., Lou, W., Bao, Y., Qiao, L., Hu, Y., Liu, K., Chen, J., Bao, D., Ye, M., Fang, Z., Gong, C., & Zhang, X. (2021). KIF18b-dependent hypomethylation of PARPBP gene promoter enhances oxaliplatin resistance in colorectal cancer. Experimental cell research, 407(2), 112827. https://doi.org/10.1016/j.yexcr.2021.112827
-
Fan, A. H., Zhao, X., Liu, H., Li, D., Guo, T., Zhang, J., Duan, L., Cheng, H., Nie, Y., Fan, D., Zhao, X., & Lu, Y. (2023). eEF1A1 promotes colorectal cancer progression and predicts poor prognosis of patients. Cancer medicine, 12(1), 513–524. https://doi.org/10.1002/cam4.4848
-
Darin Salloum, Kamini Singh, Natalie R. Davidson, Linlin Cao, David Kuo, Viraj R. Sanghvi, Man Jiang, Maria Tello Lafoz, Agnes Viale, Gunnar Ratsch, Hans-Guido Wendel; A Rapid Translational Immune Response Program in CD8 Memory T Lymphocytes. J Immunol 15 September 2022; 209 (6): 1189–1199. https://doi.org/10.4049/jimmunol.2100537
-
Rad S M, A. H., Halpin, J. C., Mollaei, M., Smith Bell, S. W. J., Hirankarn, N., & McLellan, A. D. (2021). Metabolic and Mitochondrial Functioning in Chimeric Antigen Receptor (CAR)-T Cells. Cancers, 13(6), 1229. https://doi.org/10.3390/cancers13061229
-
Yongjie Wei, Zhenyi An, Zhongju Zou, Rhea Sumpter Jr, Minfei Su, Xiao Zang, Sangita Sinha, Matthias Gaestel, Beth Levine (2015) The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation eLife 4:e05289 https://doi.org/
-
Willem A. Hanekom,Chapter 18 - Tuberculosis Vaccines,Editor(s): Barry R. Bloom, Paul-Henri Lambert,The Vaccine Book (Second Edition),Academic Press,2016,Pages 363-383,ISBN 9780128021743, https://doi.org/10.1016/B978-0-12-802174-3.00018-7
-
Wang, T., Huang, C., Lopez-Coral, A., Slentz-Kesler, K. A., Xiao, M., Wherry, E. J., & Kaufman, R. E. (2012). K12/SECTM1, an interferon-γ regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. Journal of leukocyte biology, 91(3), 449–459. https://doi.org/10.1189/jlb.1011498
-
Wilson, C. B., & Merkenschlager, M. (2006). Chromatin structure and gene regulation in T cell development and function. Current opinion in immunology, 18(2), 143–151. https://doi.org/10.1016/j.coi.2006.01.013
-
Cámara, Y., Asin-Cayuela, J., Park, C. B., Metodiev, M. D., Shi, Y., Ruzzenente, B., Kukat, C., Habermann, B., Wibom, R., Hultenby, K., Franz, T., Erdjument-Bromage, H., Tempst, P., Hallberg, B. M., Gustafsson, C. M., & Larsson, N. G. (2011). MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell metabolism, 13(5), 527–539. https://doi.org/10.1016/j.cmet.2011.04.002
-
O’Sullivan, et al (n.d.). Fever supports CD8+ effector T cell responses by Pnas.org. https://www.pnas.org/doi/full/10.1073/pnas.2023752118
-
Baba, Y., Nosho, K., Shima, K., Goessling, W., Chan, A. T., Ng, K., Chan, J. A., Giovannucci, E. L., Fuchs, C. S., & Ogino, S. (2010). PTGER2 overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 19(3), 822–831. https://doi.org/10.1158/1055-9965.EPI-09-1154
-
Chen, J. H., Perry, C. J., Tsui, Y. C., Staron, M. M., Parish, I. A., Dominguez, C. X., Rosenberg, D. W., & Kaech, S. M. (2015). Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nature medicine, 21(4), 327–334. https://doi.org/10.1038/nm.3831
-
Eberle, A. B., Lykke-Andersen, S., Mühlemann, O., & Jensen, T. H. (2009). SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nature structural & molecular biology, 16(1), 49–55. https://doi.org/10.1038/nsmb.1530
-
El-Bchiri, J., Guilloux, A., Dartigues, P., Loire, E., Mercier, D., Buhard, O., Sobhani, I., de la Grange, P., Auboeuf, D., Praz, F., Fléjou, J. F., & Duval, A. (2008). Nonsense-mediated mRNA decay impacts MSI-driven carcinogenesis and anti-tumor immunity in colorectal cancers. PloS one, 3(7), e2583. https://doi.org/10.1371/journal.pone.0002583
-
Shaolan Qin, Yong Zhou, Jianjun Chen, Yang luo, Yier Qiu, Shuiping Tu, Ming Zhong, Low levels of TSC22 enhance tumorigenesis by inducing cell proliferation in colorectal cancer, Biochemical and Biophysical Research Communications,Volume 497, Issue 4,2018,Pages 1062-1067,ISSN 0006-291X, https://doi.org/10.1016/j.bbrc.2018.02.1777777
-
Zullo, K. M., Douglas, B., Maloney, N. M., Ji, Y., Wei, Y., Herbine, K., Cohen, R., Pastore, C., Cramer, Z., Wang, X., Wei, W., Somsouk, M., Hung, L. Y., Lengner, C., Kohanski, M. H., Cohen, N. A., & Herbert, D. R. (2021). LINGO3 regulates mucosal tissue regeneration and promotes TFF2 dependent recovery from colitis. Scandinavian journal of gastroenterology, 56(7), 791–805. https://doi.org/10.1080/00365521.2021.1917650
-
Wu, Q., Yan, T., Chen, Y., Chang, J., Jiang, Y., Zhu, D., & Wei, Y. (2021). Integrated Analysis of Expression and Prognostic Values of Acyl-CoA Dehydrogenase short-chain in Colorectal Cancer. International journal of medical sciences, 18(16), 3631–3643. https://doi.org/10.7150/ijms.63953
-
Wu, Q., Yan, T., Chen, Y., Chang, J., Jiang, Y., Zhu, D., & Wei, Y. (2021). Integrated Analysis of Expression and Prognostic Values of Acyl-CoA Dehydrogenase short-chain in Colorectal Cancer. International journal of medical sciences, 18(16), 3631–3643. https://doi.org/10.7150/ijms.63953
-
Cromer, Anne & Zambrano, Alberto & R, Millon & J, Abecassis & Wasylyk, Bohdan. (2011). NUDCD2 is overexpressed in head and neck squamous cell carcinoma and is involved in nuclear division. Journal of Biochemistry and Molecular Biology In The Postgenomic Era. 1. 29.
-
Huang, J., Pan, H., Wang, J. et al. Unfolded protein response in colorectal cancer. Cell Biosci 11, 26 (2021). https://doi.org/10.1186/s13578-021-00538-z
-
Macarulla, T., Montagut, C., Sánchez-Martin, F. J., Granja, M., Verdaguer, H., Sastre, J., & Tabernero, J. (2020). The role of PIGF blockade in the treatment of colorectal cancer: overcoming the pitfalls. Expert opinion on biological therapy, 20(1), 15–22. https://doi.org/10.1080/14712598.2020.1677603
-
Newell, L. F., & Holtan, S. G. (2017). Placental growth factor: What hematologists need to know. Blood reviews, 31(1), 57–62. https://doi.org/10.1016/j.blre.2016.08.004
-
Chen, C., Liu, S., Qu, R., & Li, B. (2020). Recurrent Neoantigens in Colorectal Cancer as Potential Immunotherapy Targets. BioMed research international, 2020, 2861240. https://doi.org/10.1155/2020/2861240
-
Fattahi, F., Kiani, J., Alemrajabi, M., Soroush, A., Naseri, M., Najafi, M., & Madjd, Z. (2021). Overexpression of DDIT4 and TPTEP1 are associated with metastasis and advanced stages in colorectal cancer patients: a study utilizing bioinformatics prediction and experimental validation. Cancer cell international, 21(1), 303. https://doi.org/10.1186/s12935-021-02002-x
-
Ciafrè, S. A., Russo, M., Michienzi, A., & Galardi, S. (2023). Long Noncoding RNAs and Cancer Stem Cells: Dangerous Liaisons Managing Cancer. International journal of molecular sciences, 24(3), 1828. https://doi.org/10.3390/ijms24031828
-
Dipika Gupta, Christopher D. Heinen,The mismatch repair-dependent DNA damage response: Mechanisms and implications,DNA Repair,Volume 78,2019,Pages 60-69,ISSN 1568-7864, https://doi.org/10.1016/j.dnarep.2019.03.009
-
Kulis, M., & Esteller, M. (2010). DNA methylation and cancer. Advances in genetics, 70, 27–56. https://doi.org/10.1016/B978-0-12-380866-0.60002-2
-
Copeland, R., Solomon, M. & Richon, V. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 8, 724–732 (2009). https://doi.org/10.1038/nrd2974
-
Zhang, Z., Butler, R., Koestler, D.C. et al. Comparative analysis of the DNA methylation landscape in CD4, CD8, and B memory lineages. Clin Epigenet 14, 173 (2022). https://doi.org/10.1186/s13148-022-01399-0
-
Salz, L., Seitz, A., Schäfer, D. et al. Culture expansion of CAR T cells results in aberrant DNA methylation that is associated with adverse clinical outcome. Leukemia 37, 1868–1878 (2023). https://doi.org/10.1038/s41375-023-01966-1
-
Yongjie Wei, Zhenyi An, Zhongju Zou, Rhea Sumpter Jr, Minfei Su, Xiao Zang, Sangita Sinha, Matthias Gaestel, Beth Levine (2015) The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation eLife 4:e05289 https://doi.org/
-
Darin Salloum, Kamini Singh, Natalie R. Davidson, Linlin Cao, David Kuo, Viraj R. Sanghvi, Man Jiang, Maria Tello Lafoz, Agnes Viale, Gunnar Ratsch, Hans-Guido Wendel; A Rapid Translational Immune Response Program in CD8 Memory T Lymphocytes. J Immunol 15 September 2022; 209 (6): 1189–1199. https://doi.org/10.4049/jimmunol.2100537
-
Ping Luo, Xiaoyan Liu, Zehai Tang & Bei Xiong (2022) Decreased expression of HBA1 and HBB genes in acute myeloid leukemia patients and their inhibitory effects on growth of K562 cells, Hematology, 27:1, 1003-1009, DOI: 10.1080/16078454.2022.2117186
-
Wilson, C. B., & Merkenschlager, M. (2006). Chromatin structure and gene regulation in T cell development and function. Current opinion in immunology, 18(2), 143–151. https://doi.org/10.1016/j.coi.2006.01.013
-
Torre, D., Fstkchyan, Y. S., Ho, J. S. Y., Cheon, Y., Patel, R. S., Degrace, E. J., Mzoughi, S., Schwarz, M., Mohammed, K., Seo, J. S., Romero-Bueno, R., Demircioglu, D., Hasson, D., Tang, W., Mahajani, S. U., Campisi, L., Zheng, S., Song, W. S., Wang, Y. C., Shah, H., … Marazzi, I. (2023). Nuclear RNA catabolism controls endogenous retroviruses, gene expression asymmetry, and dedifferentiation. Molecular cell, 83(23), 4255–4271.e9. https://doi.org/10.1016/j.molcel.2023.10.036
-
Vibha Rani,Chapter 11 - microRNAs as critical regulators in heart development and diseases,Editor(s): Buddhi Prakash Jain, Shyamal K. Goswami, Tapan Sharma,In Translational Epigenetics,Post-Transcriptional Gene Regulation in Human Disease,Academic Press,Volume 32,2022,Pages 187-203,ISBN 9780323913058, https://doi.org/10.1016/B978-0-323-91305-8.00005-3. (https://www.sciencedirect.com/science/article/pii/B9780323913058000053)
-
FOXO1-regulated lncRNA CYP1B1-AS1 suppresses breast cancer cell proliferation by inhibiting neddylation - Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Upregulation-of-CYP1B1-AS1-inhibits-cell-proliferation-and-induces-apoptosis-a_fig3_373480558
-
Zhou, L., Jiang, Y., Luo, Q. et al. Neddylation: a novel modulator of the tumor microenvironment. Mol Cancer 18, 77 (2019). https://doi.org/10.1186/s12943-019-0979-1
-
Wang, L. N., Gao, M. H., Wang, B., Cong, B. B., & Zhang, S. C. (2018). A role for GPI-CD59 in promoting T-cell signal transduction via LAT. Oncology letters, 15(4), 4873–4881. https://doi.org/10.3892/ol.2018.7908
-
Higashi, S., Iseki, E., Minegishi, M., Togo, T., Kabuta, T., & Wada, K. (2010). GIGYF2 is present in endosomal compartments in the mammalian brains and enhances IGF-1-induced ERK1/2 activation. Journal of neurochemistry, 115(2), 423–437. https://doi.org/10.1111/j.1471-4159.2010.06930.x
-
Andreas Wyttenbach, Jina Swartz, Hiroko Kita, Thomas Thykjaer, Jenny Carmichael, Jane Bradley, Rosemary Brown, Michelle Maxwell, Anthony Schapira, Torben F. Orntoft, Kikuya Kato, David C. Rubinsztein, Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease, Human Molecular Genetics, Volume 10, Issue 17, 15 August 2001, Pages 1829–1845, https://doi.org/10.1093/hmg/10.17.1829
-
Li, F., Wang, L., Wang, Y., Shen, H., Kou, Q., Shen, C., Xu, X., Zhang, Y., & Zhang, J. (2023). HECW2 promotes the progression and chemoresistance of colorectal cancer via AKT/mTOR signaling activation by mediating the ubiquitin-proteasome degradation of lamin B1. Journal of Cancer, 14(15), 2820–2832. https://doi.org/10.7150/jca.87545
-
Lämmerhirt, L., Kappelmann-Fenzl, M., Fischer, S., Pommer, M., Zimmermann, T., Kluge, V., Matthies, A., Kuphal, S., & Bosserhoff, A. K. (2022). Knockdown of Lamin B1 and the Corresponding Lamin B Receptor Leads to Changes in Heterochromatin State and Senescence Induction in Malignant Melanoma. Cells, 11(14), 2154. https://doi.org/10.3390/cells11142154
-
Narayanankutty A. (2019). PI3K/ Akt/ mTOR Pathway as a Therapeutic Target for Colorectal Cancer: A Review of Preclinical and Clinical Evidence. Current drug targets, 20(12), 1217–1226. https://doi.org/10.2174/1389450120666190618123846
-
Li, F., Wang, L., Wang, Y., Shen, H., Kou, Q., Shen, C., Xu, X., Zhang, Y., & Zhang, J. (2023). HECW2 promotes the progression and chemoresistance of colorectal cancer via AKT/mTOR signaling activation by mediating the ubiquitin-proteasome degradation of lamin B1. Journal of Cancer, 14(15), 2820–2832. https://doi.org/10.7150/jca.87545
-
Vidhya Krishnamoorthy, Richa Khanna, Veena K. Parnaik,E3 ubiquitin ligase HECW2 targets PCNA and lamin B1,Biochimica et Biophysica Acta (BBA) - Molecular Cell Research,Volume 1865, Issue 8,2018,Pages 1088-1104,ISSN 0167-4889, https://doi.org/10.1016/j.bbamcr.2018.05.008.
-
Li, J., Tian, X., Nie, Y., He, Y., Wu, W., Lei, X., Zhang, T., Wang, Y., Mao, Z., Zhang, H., Zhang, X., & Song, W. (2021, December 3). BTBD10 is a prognostic biomarker correlated with immune infiltration in hepatocellular carcinoma. Frontiers. https://www.frontiersin.org/articles/10.3389/fmolb.2021.762541/full
-
Nawa, M., & Matsuoka, M. (2013). KCTD20, a relative of BTBD10, is a positive regulator of Akt. BMC biochemistry, 14, 27. https://doi.org/10.1186/1471-2091-14-27
-
Huan, T., Chen, D., Liu, G., Zhang, H., Wang, X., Wu, Z., Wu, Y., Xu, Q., & Yu, F. (2022, January 15). Activation-induced cell death in car-T cell therapy - human cell. SpringerLink. https://link.springer.com/article/10.1007/s13577-022-00670-z
-
Uhlen M et al., A pathology atlas of the human cancer transcriptome. Science. (2017)
PubMed: 28818916 DOI: 10.1126/science.aan2507
-
R. Fuentes, N. R. R. (n.d.). Membrane therapy using DHA suppresses epidermal growth factor ... Journal of Lipid Research. https://www.jlr.org/article/S0022-2275(21)00006-7/fulltext
-
Tan, L., Fu, D., Liu, F., Liu, J., Zhang, Y., Li, X., Gao, J., Tao, K., Wang, G., Wang, L., & Wang, Z. (2023). MXRA8 is an immune-relative prognostic biomarker associated with metastasis and CD8+ T cell infiltration in colorectal cancer. Frontiers in oncology, 12, 1094612. https://doi.org/10.3389/fonc.2022.1094612
-
Valieva Y, Ivanova E, Fayzullin A, Kurkov A, Igrunkova A. Senescence-Associated β-Galactosidase Detection in Pathology. Diagnostics. 2022; 12(10):2309. https://doi.org/10.3390/diagnostics12102309
Acknowledgement
I would like to thank Renauer, P., Park, J. J., Bai, M., Acosta, A., Lee, W. H., Lin, G. H., Zhang, Y., Dai, X., Wang, G., Errami, Y., Wu, T., Clark, P., Ye, L., Yang, Q., & Chen, S., at Yale's Genetics Research Lab, for publishing the raw transcriptomes utilized in this study. I appreciate their strides in assuring science is as accessible as possible, and their indirect involvement in advancing youth research and the academic scene as a whole.
I would also like to thank my science fair coordinator, Ms. Bretner. Thank you for working so hard to manage everything and reassuring us every step of the way!
Lastly, I would like to thank my family, friends, and specifically: Kriti, Maryam, Swathi, Yasmine, Nishi, Bronnie, Ruqayyah, Nikole, Rianna, Angelina, and Arushi for the reassurance. Sorry if I forgot anyone!
Thank you to all my science fair friends too, for being there to bounce ideas, motivate, and critique my methods.